Figures & data
Figure 1 MiR biology. (A) MiR production. MiRs occur inter- and intragenically and can be transcribed by RNA Polymerase II or III (Pol-II or Pol-III).Citation23 Prior to nuclear export, the “pri-miR” hairpin is excised from the initial transcript by Drosha. Following nuclear export, the hairpin is processed by Dicer to produce the ∼20 nt mature miR. Image adapted from Bartel et al.Citation54 (B) MiR seeds and seed matches. Cartoon depicting a perfect seed match between a mature miR (top) and a target mRNA (bottom). The miR nucleotides commonly referred to as a “seed” (basepairs 2 through 8) and a perfect seed match in the mRNA are shown in red. Vertical lines indicate basepairing.
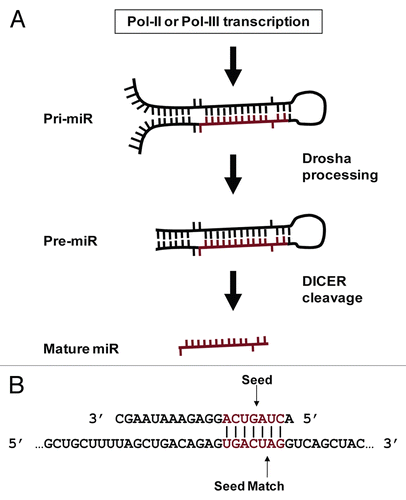
Figure 2 MiRs commonly occur at the intersection of related, converging TEs. (A) Cartoon depicting the theoretical origin of numerous miRs. A pri-miR is depicted just above an arrow indicating read through transcription from a positive strand LINE1 (L1) element into an adjacent negative strand L1. This relationship suggests a likely series of events leading to the creation of a potential miR hairpin in which an L1 is inserted immediately adjacent to a related L1 on the opposite strand creating the convergent or “tail to tail” organization illustrated. Next, transcriptional read through would result in an imperfect RNA hairpin being produced potentially recognized and processed by the RNAi machinery with each stem corresponding to the terminal nucleotides of the contributing LINEs. (B) Examples of human miR loci alignments to the RepBase dataset. Importantly, all pre-miRs significantly aligning with a Censor Server repetitive element annotation have been reported irrespective of agreement with the scenario portrayed in (A)—while we find numerous loci arising by this mechanism, we find others (like miR-640) do not. Entirely contained within an THER1 SINE, we propose an additional mechanism (point mutation(s) resulting in an alteration of normal SINE secondary structure gave rise to pre-miR-640). All repetitive elements (grey rectangles) occurring within 500 bp (5′ and 3′) have been included in the scale diagrams for uniformity. The RepBase repetitive element annotations found in these diagrams are described immediately beneath each locus as “Element 1, Element 2, etc.,” as they occur 5′ to 3′. “Base Positions” refers to the basepairs occupied by a miR hairpin (in the current Ensembl assembly). All loci have been diagrammed with respect to the Watson strand and the orientation of internal elements indicated by position above (5′ to 3′) or below (3′ to 5′) the center line. Element basepair positions are in respect to distance (±) from the 1st nucleotide of the pre-miR (as occurring on the Watson strand). *previously described origin.Citation21,Citation23 Figures adapted from references Citation21 and Citation23.
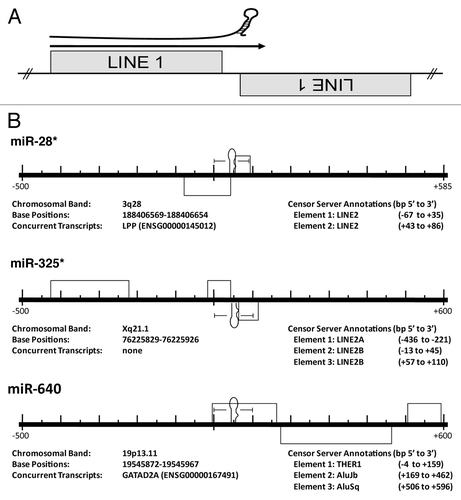
Figure 3 MiR-284 familial alignment. Alignment of the 12 miR-284 hairpins. Individual hairpin sequences along with species (right) and miRBase identifier (left) are shown. *indicates 100% nucleotide conservation. Grey highlight indicates specific miR hairpins annotated as bearing significant sequence complementarity to Mermite-35.

Figure 4 MiR-28 alignments with predicted targets. Alignments between three predicted miR-28 target 3′UTRs (top), a consensus L2 LINE (middle) and the miR-28 genomic sequence reverse complemented (bottom) are illustrated. Mature miR-28 is highlighted in grey. Open boxes indicate perfect seed matches. To qualify as a 3′UTR “hit”, alignments were required to (1) contain a perfect seed match, (2) match .50% of the flanking sequence used in the target query and (3) occur within a 3′UTR sequence annotated as an L2 sequence by Censor Server. Vertical lines indicate base identity with the L2 consensus sequence. Dotted lines indicate purine/pyrimidine conservation. LYPD3, “LY6/PLAUR domain containing 3”; E2F6, “E2F transcription factor 6”; CXCL9, “chemokine (C-X-C motif) ligand 9”.
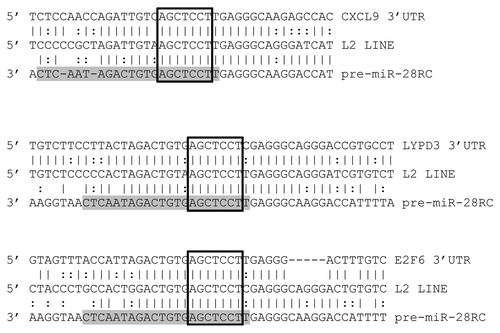
Figure 5 Molecular events responsible for miR establishment. MiRs “arise” when an advantageous regulatory niche has developed out of a series of random TE insertions after which the fortuitous formation of a TE juxtaposition (shown, ) and subsequent processing by RISC can lead to miR establishment if the resulting small RNAs confer some regulatory advantage in order to be selected for (e.g., improved cell tolerance to apoptotic stimulus due to delayed response accompanying translational repression). Numbers indicate the sequential steps necessary for miR establishment. Thick lines indicate genomic DNA and thin lines denote RNA.
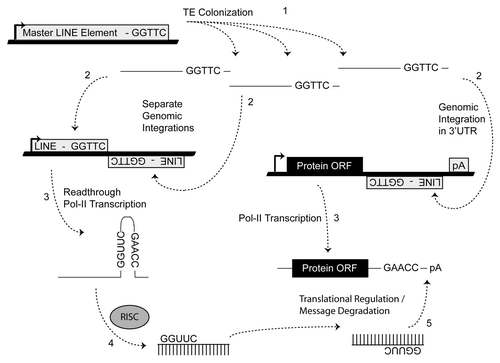
Table 1 Summary of MiR loci progenitor transposable elements
Table 2 Summary of familial inclusions