Figures & data
Figure 1 Models of the retroelement retrotransposition cycle. (Left part) Retrotransposition pathway of endogenous retroviruses (or LTR retrotransposons) such as murine IAP and MusD. Mammalian endogenous retroviruses sequences are structurally similar to infectious retroviruses such as HIV-1 and MLV. The infectious retroviruses encode an envelope protein (Env) that facilitates their transmission from one cell to another, whereas endogenous retroviruses either lack or contain a remnant of an env gene and can only integrate into the genome at a new site within their cell of origin. Endogenous retroviruses also contain slightly overlapping ORFs for their group-specific antigen (Gag), protease (Prt), polymerase (Pol) and terminal LTRs. The pol genes encode a reverse transcriptase, ribonuclease H and integrase to provide enzymatic activities for generating proviral cDNA from viral genomic RNA and inserting it into the host genome. Their life cycle includes the formation of VLPs, which remain intracellular. Reverse transcription of endogenous retrovirus genomic RNA occurs within the VLP in the cytoplasm, and is a complicated, multistep process. (Right part) Retrotransposition pathway of L1 retroelements. A ∼6 kb functional L1 element contains an internal RNA polymerase II promoter in the 5′ UTR , followed by two open reading frames. ORF1 encodes RNA-binding protein (ORF1p) that is required for ribonucleoprotein particle (RNP) formation in the cytoplasm. ORF2 encodes a protein with endonuclease and reverse transcriptase activities (ORF2p). Both ORF1p and ORF2p are critical for retrotransposition by a “copy and paste” mechanism. A short 3′ UTR is followed by a poly(A) tail, and the entire element is flanked by target site duplications (TSD). L1 DNA synthesis in the nucleus is based on “target-primed reverse transcription (TPRT)” in which ORF2p nicks target chromosomal DNA, using the resultant 3′-OH to prime the reverse transcription of L1 RNA as a template. At an early phase of replication, L1 RNA forms a RN P complex in the cytoplasm as a retrotransposition intermediate by associating with ORF1p and ORF2p.
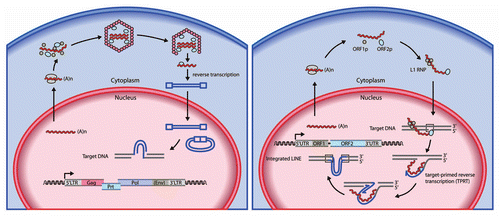
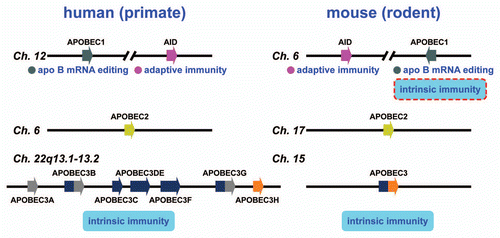
Figure 2 AID/APOBEC genes in human and murine genomes. A schematic of the human and murine genomes containing members of the AID/APOBEC family. There are several forms of mammalian AID/APOBEC proteins with distinct functions in vivo: activation-induced deaminase (AID), APOBEC1(A1), APOBEC2(A2), APOBEC3(A3) and APOBEC4(A4). The primate-specific cluster of at least seven A3 related genes: A3A-3H, resides on the same chromosome within ∼150 kb. In contrast, mice retains only a single A3 gene, located on chromosome 15e2, indicating a relatively recent, and possibly unprecedented gene expansion has occurred in mammalian species. Each A3 gene encodes a protein with one or two conserved zinc-coordinating motifs (Z1, Z2 or Z3).Citation25,Citation30 Grey and blue denote Z1 and Z2, respectively, while orange denotes Z3. Rodents encode only one A3 gene (a Z2–Z3 fusion). A1 and AID are located approximately 900 kb apart on human chromosome 12.