Figures & data
Figure 1. MiR biology and origins. (A) MiR generation. MiRs can occur inter- or intragenically and be transcribed by either RNA Polymerase II or III.Citation24 Following transcription, the “pre-miR” hairpin (middle) is excised from the initial transcript (or pri-miR) (top) by Drosha. Once in the cytoplasm, the hairpin or stem loop is cleaved and denatured by Dicer to excise the ~20 nt mature miR (bottom). (B) MiR seeds. A seed match between a miR (top) and target mRNA (bottom) is illustrated. The nucleotides in a miR generally referred to as a “seed” (nts 2 through 8) and a “seed match” in a mRNA are depicted in red. Basepairing is indicated by vertical lines. (C) Cartoon depicting the molecular origin of many miR loci. MiRs were initially formed by the neighboring insertions of related TEs. A pri-miR is depicted just above the genome with an arrow indicating readthrough Pol-III transcription from a (+) strand Alu SINE into a neighboring (-) strand Alu. As illustrated, transcriptional readthrough would generate a RNA stem loop whose stems (loaded into the RISC machinery if processed) would correspond to the terminal nucleotides of the neighboring Alus. Figure adapted from.Citation23
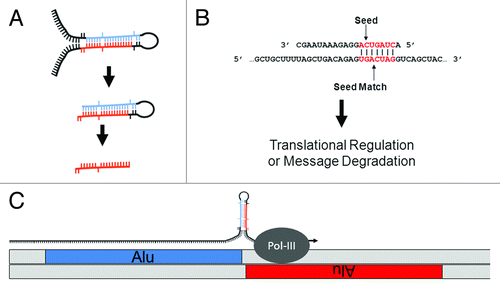
Figure 2. Establishing a miR regulatory network. MiR regulatory networks are formed when an advantageous regulation arises from a series of random TE insertions into expressed genomic loci, and the formation of a TE juxtaposition by the positive and negative strand insertions of related TEs. Thick lines indicate genomic DNA and thin lines denote RNA. Figure adapted from.Citation23
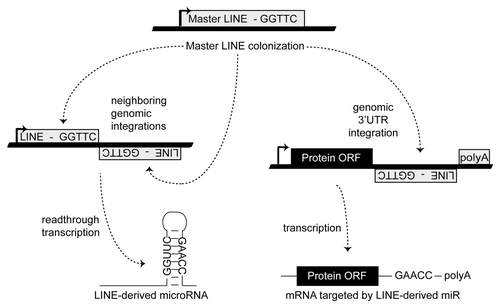
Table 1. OrbId summary. The full EnsemblCitation36 set of 178,375 unique human mRNA transcripts including 5′UTR, 3′UTR, and ORF annotations were compiled in and retrieved using the Biomart mining utilityCitation37. “Human miRs analyzed” correspond to the full set of human miR mature sequences identified by Borchert et al. as originating from TEsCitation23 and were obtained from the miR Registry miRBase.Citation2
Figure 3. MiR-28 predicted target three way alignments. Alignments between OrbId predicted miR-28 target mRNAs (middle), a consensus L2B LINE (L2Plat1o) (top), and miR-28 (bottom). (*), base identity in the three aligning sequences. (^), base identity (indicating base pairing) between the miR and mRNA target only. (:), GU basepairing between miR and mRNA target. 3′ UTR or 5′ UTR targeting is indicated. Uracils are shown as thymines and UTRs have been reverse complemented for illustrative purposes.

Table 2. OrbId prediction set for select TE-derived human miRs. “miR Name” refers to miRBaseCitation2 annotation while “Ensembl Gene ID” and “Gene Name” were obtained using the Biomart mining utilityCitation37. “Diana, TS” refers to whether a predicted target is contained within publically accessible Diana (D) and TargetScan (TS) predictionsCitation17,Citation21. “Region” refers to the location of a predicted target site within a given mRNA. MiR-28–5p corresponds to the participating member of the miR-28 family. MiR-1254–1 is a member of the Alu-miR family. MiR-603 is a member of the miR-548 family
Figure 4. MiR-28, miR-151 and miR-708 target network. Only shared targets are depicted including 14 of 15 miR-28–5p targets, 11 of 14 miR-151a-5p targets, and 4 of 13 miR-708 targets. Green lines indicate miR regulation.
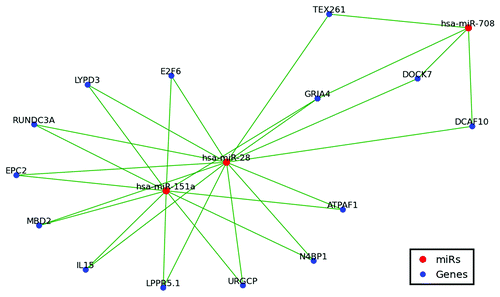
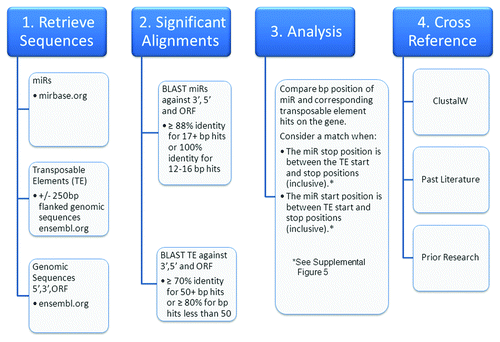
Figure 5. OrbId methodology flowchart. A high level overview of the steps taken to determine miR and transposable element concurrent alignments within the human transcriptome.