Figures & data
Figure 1. Shared 3′ end of paired autonomous and non-autonomous elements. Non-LTR elements and their non-autonomous partners (LINE/SINE pairs) can be subdivided into two subgroups based on their shared 3′ end sequence: (A) stringent and (B) relaxed, reviewed in.Citation50 Schematics of representative examples of the two subgroups are shown. The “stringent” LINE/SINE pair group (A) share the 3′ end sequence (green box). Several different LINE/SINE pairs from reptiles, fish, mammals and plants fall into this category. SINEs in this subgroup derived their 3′ end from the LINE sequence. A stem-loop structure in the 3′end is commonly observed in these LINE/SINE pairs (shown as a loop). The transcripts of the “relaxed” SINE/LINE pair group (B) share a 3′-poly(A) tail, which include the human LINE-1 element with its two open reading frames, ORF1 (purple) and ORF2 (orange). LINE-1 provides the proteins that mobilize several non-autonomous RNAs in trans: mRNAs generating processed pseudogenes and the composite retrotransposon SVA (SINE-VNTR-Alu) element which are transcribed by the RNA polymerase II (pol II); and the RNA polymerase III (pol III) transcribed SINEs that are ancestrally derived from pol III genes, such as 7SL and tRNAs. Some of these SINE transcripts form secondary structures (represented as looped lines inside the boxes) that are important for retrotransposition. One example is Alu (shown as a dimeric molecule where each non-identical side was individually derived from the 7SL gene, represented as blue boxes separated by an A-rich region represented by “AA”). “U”s at the end of the transcripts represent pol III termination and blue “N”s represent sequences preceding the pol III termination signal derived from the flanking DNA of the specific locus generating the transcript. However, the vast majority of SINEs are derived from tRNA genes (represented as a purple box). The SINEs with polyA tails can be further subdivided into two classes, class T- and class T+, depending on the presence/absence of a polyadenylation signal (pAS) and a TC-motif preceding the pol III terminator and polyA tail at the 3′ end.Citation29
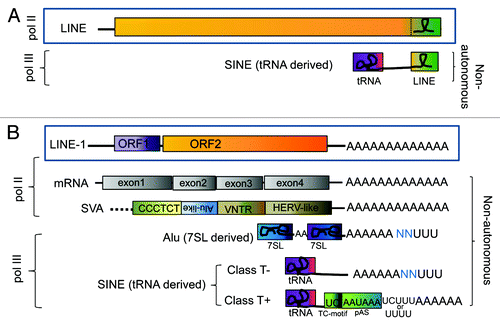
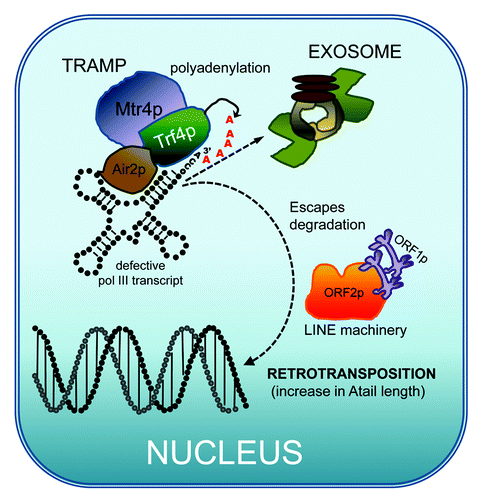
Figure 2. SINE genesis model. RNA Pol III products such as tRNAs, U6, 5S rRNA and 7SL undergo nuclear surveillance. Defective or incorrectly formed pol III transcripts are targeted for destruction in the nucleus by the exosome (reviewed in ref. Citation40). One of the steps in the surveillance of the pol III RNAs occurs through the polyadenylation of the transcript by the TRAMP complex. The diagram shows TRAMP4 composed of Rrf4p, Mtr4p and Air2p in humans. It is proposed that the addition of the polyA tail at the 3′ end signals the exosome to degrade the RNA. In our model, we propose that a potential outcome is that a few of the defective polyadenylated tRNAs or other pol III RNAs escape degradation by the exosome. This will in turn allow the stochastic interaction with the LINE retrotransposition machinery (in primate lineage expect to find L1 ORF2 protein and L1 ORF1 proteins) already present in the nucleus. LINE proteins may vary in other lineages, such as plants. The retrotransposition of the polyadenylated pol III transcript into the DNA would generate the first ancestral element. Fortuitous integration in a transcriptionally supportive site could lead to the creation of an active element, i.e., the birth of a SINE. In this model, the expansion of the A-tail during insertion caused by the LINE ORF2 protein reverse transcription slippage would significantly favor the introduction of the critical A-tail length to generate a retrotranspositionally competent insert.