Figures & data
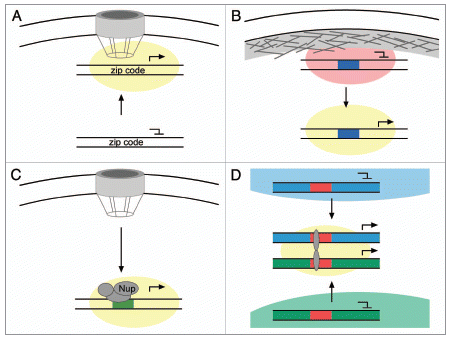
Figure 1 Gene relocalization within the nucleus associated with changes in expression. (A) Genes in yeast, and perhaps other organisms, associate with the nuclear pore complex upon activation. Interaction of the INO1 and TSA2 genes with the NPC requires DNA zip codes in their promoters and promotes their expression, perhaps because of a high local concentration of factors that promote transcription (yellow cloud). (B) Genes that are induced during development often relocalize away from the nuclear periphery upon activation. Interaction with the nuclear lamina is associated with transcriptional repression (red cloud). It is possible that cis-acting DNA elements within such genes (blue box) might either target them to the nuclear lamina when repressed or target them to an internal site when activated. (C) Some Drosophila genes interact with NPC proteins such as Nup153, Nup98, Sec13 and Mtor in the nucleoplasm and this promotes their expression. It is possible that cis-acting DNA elements (green box) control the interaction with NPC proteins specifically within the nucleoplasm. (D) Co-regulated genes from different chromosomes can cluster together upon activation, a phenomenon known as “gene kissing”. These genes often relocalize from within chromosome territories to the inter-chromosomal space. Colocalization of genes at “transcription factories” requires transcriptional regulators (grey ovals) that bind to cis-acting DNA elements (red boxes).