Figures & data
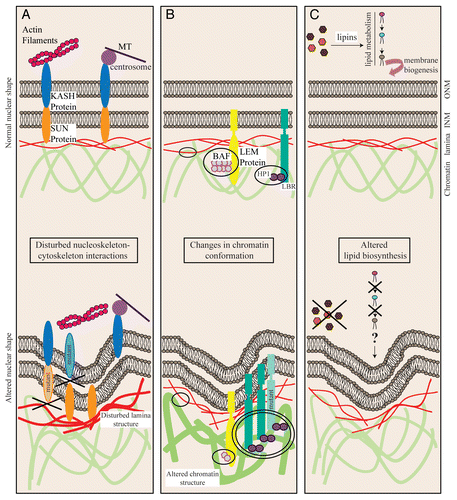
Figure 1 The farnesylated nuclear membrane protein Kuk affects nuclear shape in different cell types. (A) Surface view of a wt Drosophila embryo (left) and of an embryo with six genomic copies of kuk (right), in late cellularization. Kuk is used as a marker of the NM. Scale bar: 10 µm. (B) Nuclear morphology upon Kuk overexpression in NIH-3T3 cells. The nucleus of a transiently transfected NIH-3T3 cell expressing Kuk (green) is shown together with the nucleus of a non transfected cell. Lamin A/C (red) marks the NM. Scale bar: 5 µm. (C) Nuclear morphology of control (left) and GFP-Kuk expressing (right) S. cerevisiae in mid-log phase. mCherry-Nup133 (red) marks the NM and GFP-Kuk is shown in green. Scale bar: 2.5 µm.
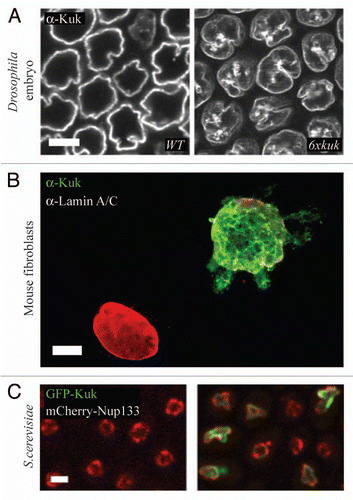
Figure 2 Farnesylated nuclear membrane proteins affect nuclear shape by directly interacting with the lipid bilayer. Schematic representation of our current model, explaining how farnesylated proteins of the nuclear membrane affect nuclear shape by directly interacting with the lipid bilayer of the INM via their farnesylated C-terminus. The arrows indicate sites where the C-terminal part of the protein is interacting with the lipid bilayer.
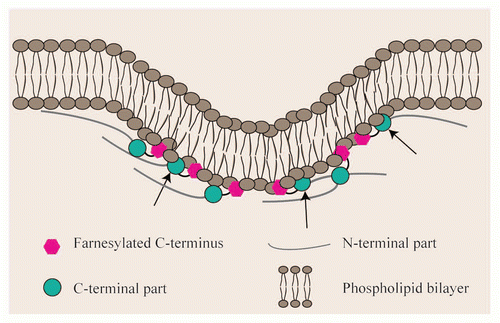
Figure 3 Cytoskeletal elements, chromatin structure and lipid biosynthesis are involved in shaping the nuclear membrane. (A) Disturbed nucleoskeleton-cytoskeleton interactions may be caused i.e., by modified levels of SUN or KASH proteins, by mutations in KASH or SUN proteins or by abnormal lamina structure. In all cases, the interaction of lamina-INM-ONM-cytoskeleton is not properly established. (B) Interactions of the lamina with chromatin (in circles) are either direct or mediated by the interaction of INM proteins with chromatin binding proteins i.e., LEM proteins with BAFCitation46,Citation57 or LBR with HP1.Citation58 Disturbed interaction of lamina and chromatin either by general changes in chromatin conformation or by modified levels of the proteins mediating the interaction might lead in changes in nuclear shape. The chromatin lamina interaction might be lost (circles) or enhanced i.e., due to increased LBR amounts or to mutations in LBR (double circle). (C) Altered lipid biosynthesis i.e., by mutations in lipins is suggested to influence membrane structure, even though the mechanism remains unclear.