Figures & data
Figure 1A (See next page for B–E). DamID of Ima1 and Man1. (A) Subnuclear localization of Ima1- and Man1-Dam fusion proteins and the Dam-only control. Immunofluorescence microscopy was performed using an anti-myc antibody against the 7myc-tags in the expressed proteins. A FITC-coupled antibody was used for secondary staining.
Figure 1B–E (See previous page for A). DamID of Ima1 and Man1. . (B) Schematic drawing of the DamID method with INM-proteins. GATC sequences that come in close proximity to the nuclear envelope will be methylated by the Dam-methylase fused to the respective INM protein. Methylated DNA sequences are amplified using restriction digests and adaptor-mediated PCR. (C) Example distribution of Ima1 and Man1 over ~70 kb on chromosome 1. Regions defined as targets by the Hidden Markow Model are colored in green and blue, respectively. Genes are colored by their expression statusCitation25 when available, otherwise shown in white. (D) Target distribution of Man1 and Ima1 in the S. pombe genome. Targets were defined using a Hidden Markow Model on array signals for all probes mapping uniquely in the genome. (E) Scatterplot of Man1-Dam ID scores vs. Ima1-DamID scores. Dots are colored based on their annotation by the Hidden Markow Model (Ima1-targets, Man1-targets, both, neither).
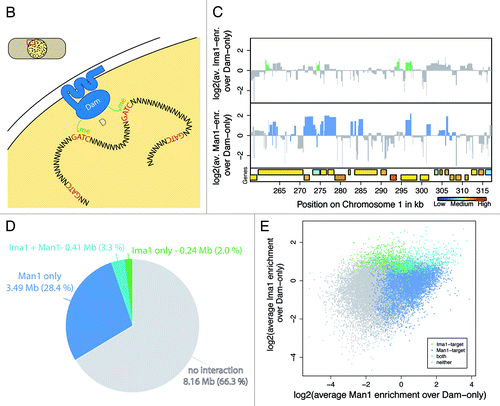
Figure 2. Peripheral genes show lower expression levels and point preferentially away from the nuclear envelope. (A) Average enrichments of Ima1 and Man1 over the body of genes with different expression levels, represented by boxplots. Only those genes for which expression data was available are displayed (4884 of 5027). p-values are calculated using Wilcoxon-Mann-Whitney tests comparing the distribution of one gene expression level to the distribution of all remaining genes. (B) Average enrichments of Ima1 and Man1 over intergenic regions (IGRs) of different orientations. P-values are calculated using Wilcoxon-Mann-Whitney tests comparing the distribution of one IGR orientation to the distribution of all remaining IGRs.
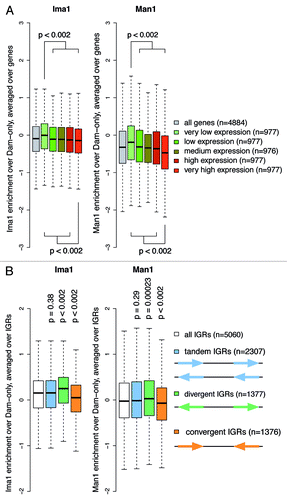
Figure 3. Peripheral chromatin shows low occupancy of RNAP II and nucleosomes, as well as increased levels of H2A.Z. Overview boxplots for (A) RNAP II occupancy,Citation50 (B) nucleosome densityCitation27 and (C) H2A.Z occupancy.Citation28 Boxplots show score distributions over target loci, depending on how they are annotated by the Hidden Markow Model. P-values are calculated by permutation test. Number of loci plotted per box and genome portion covered: genome wide, 29,697 loci (12.30 Mb); Man1 only, 6,429 loci (3.49 Mb); Ima1 only, 645 loci (0.24 Mb); both, 826 loci (0.41 Mb); neither, 21,797 loci (8.16 Mb).
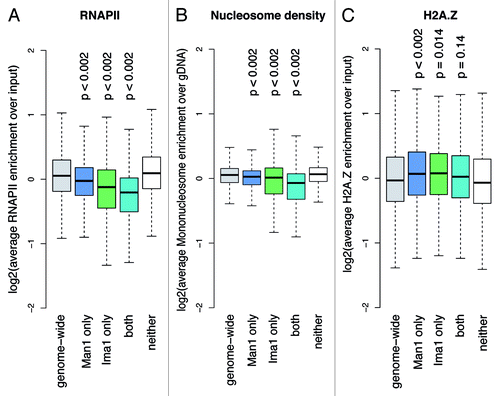
Figure 4. Ima1 shares common targets with the RNAi components Dcr1 and Rdp1. Overview boxplots for (A) Dcr1 and (B) Rdp1 occupancy.Citation32 The boxplots show score distributions over target loci, depending on how they are annotated by the Hidden Markow Model. The P-values were calculated by permutation test. Number of loci plotted per box and genome portion covered: genome wide, 29,697 loci (12.30 Mb); Man1 only, 6,429 loci (3.49 Mb); Ima1 only, 645 loci (0.24 Mb); both, 826 loci (0.41 Mb); neither, 21,797 loci (8.16 Mb).
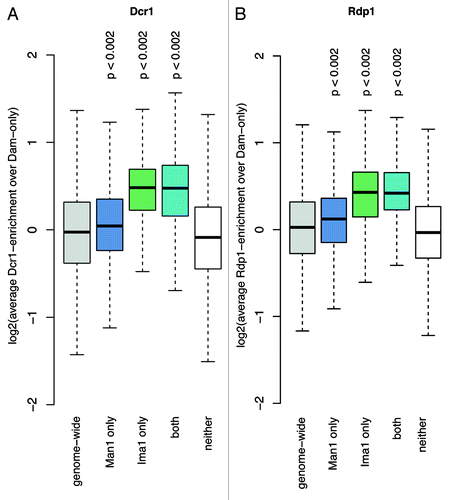
Figure 5. Man1 is enriched at sub-telomeres and binds to Swi6-targets. (A) Target distribution along sub-telomeres. Genome browser views showing the distributions of Man1, Ima1, Swi6Citation32 and H2A.ZCitation28 as log2-enrichment, as well as H3K9me2 and H3K4me2Citation51 enrichment over the sub-telomere of chromosome 2. Genes are colored by their expression statusCitation25 when available, otherwise shown in white. LTR elements are depicted as black boxes. (B) Overview boxplot for Swi6.Citation32 The boxplot shows score distributions over target loci, depending on how they are annotated by the Hidden Markow Model. P-values are calculated by permutation test. Number of loci plotted per box and genome portion covered: genome wide, 29,697 loci (12.30 Mb); Man1 only, 6,429 loci (3.49 Mb); Ima1 only, 645 loci (0.24 Mb); both, 826 loci (0.41 Mb); neither, 21,797 loci (8.16 Mb).
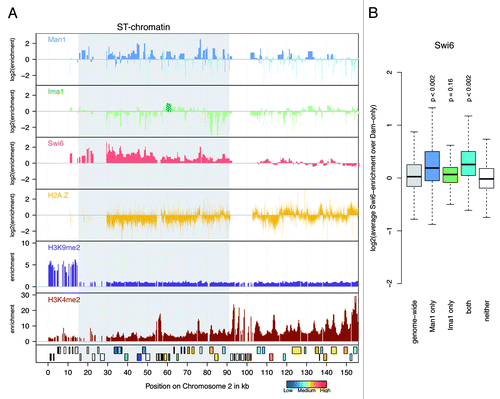
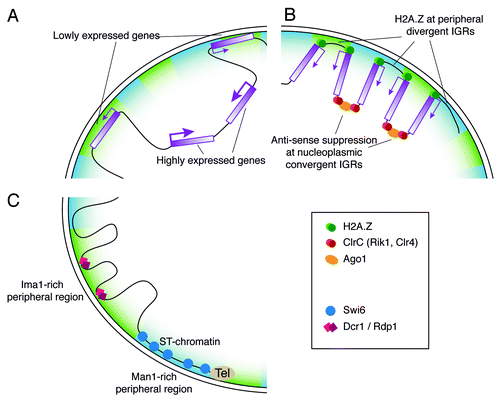
Figure 6. Putative model for chromatin organization at the fission yeast nuclear periphery. (A) Gene expression at the nuclear periphery. Genes with low expression levels are more commonly associated with the nuclear periphery, while highly expressed genes tend to reside in the nuclear interior. (B) A potential role for H2A.Z at the nuclear envelope. Localization of divergent IGRs and H2A.Z at the nuclear envelope could present a mechanism for anchoring the promoters of convergent gene pairs at the periphery. The convergent IGR would then be located in the nuclear interior, where the RNA surveillance machinery can act to suppress anti-sense transcription. (C) Differential localization of Ima1 and Man1. The INM proteins Ima1 and Man1 are not equally distributed at the nuclear periphery, but rather occupy distinct areas that interact with different chromosomal regions. The subtelomeric chromatin is associated with Man1-rich peripheral regions, where Swi6 is also located. In contrast, Ima1 is absent from subtelomeric chromatin, but tends to associate with Dcr1 and Rdp1 target loci.