Figures & data
Table 1. Current approaches used to map distal enhancers genome-wide
Figure 1. Schematic view of the epigenetic dynamics at induced genes during early T-cell differentiation. In the early CD4-CD8- double negative (DN) thymocytes, the enhancer is enriched for H3K4me1 and is found in a poised configuration. During differentiation into CD4+CD8+ double positive (DP) thymocytes, induced enhancers recruit Pol II and acquire H3K4me3. The profiles shown here are inspired of canonical loci described in reference Citation18.
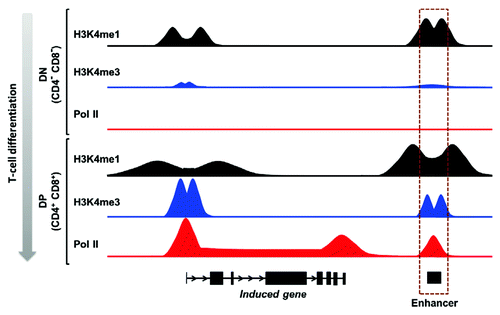
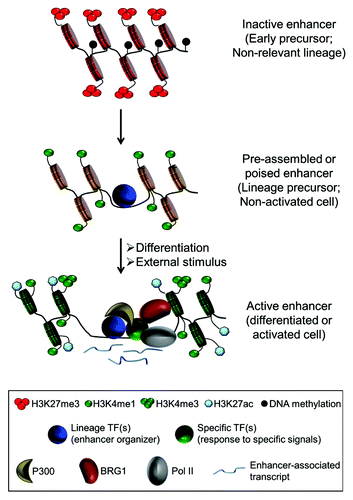
Figure 2. Chromatin dynamics at tissue-specific enhancers during cell differentiation. In early precursors or non-relevant lineages, the enhancer region is cover by nucleosomes and often associated to repressive marks, such as H3K27me3 or DNA methylation. During differentiation, lineage-specific TFs (also called master regulators or enhancer organizers) bind to the majority of the tissue-specific enhancer repertoire. These enhancers are nucleosome free regions and enriched for H3K4me1, but are generally in a poised state. Subsequently, upon cell differentiation and/or external stimuli, induced or activated TFs binds to some of the accessible enhancers in order to fine-tune gene expression. Active enhancers are now associated with additional cofactors such as BRG1, p300 and Pol II and correlate with further nucleosome remodeling, acquisition of additional histone modifications, such as H3K27ac and H3K4me3, and local transcription.