Figures & data
Figure 1 NE proteins interact with chromatin proteins. The NE consists of outer (ONM) and inner (INM) nuclear membranes that fuse where the nuclear pore complexes (NPCs) are inserted. Both ONM and INM contain membrane spanning proteins that are generally referred to as NETs for Nuclear Envelope Transmembrane protein. Underlying the INM is the lamin intermediate filament polymer (green). The INM harbors a specific set of NETs (red), which together with the lamins are referred to as the lamina. Lamins, NETs and NPCs can all interact with chromatin components like Barrier-to-Autointegration Factor (BAF), Heterochromatin Protein 1 (HP1) and histones.
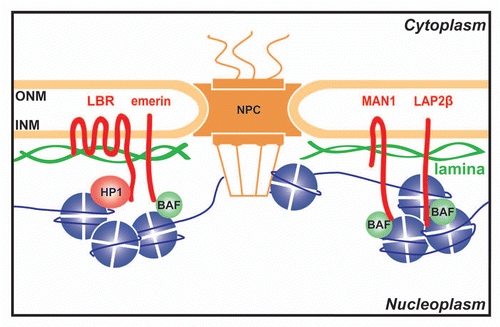
Figure 2 Each chromosome has a distinctive positioning in the nucleus with respect to the nuclear periphery. In human HT1080 fibroblast cells chromosomes 5 and 17 tend to be in the nuclear interior while chromosome 13 tends to be at the periphery. The chromosome is shown in green and the DNA from DAPI staining in blue delineates the nuclear boundary. Cells were fixed with formaldehyde prior to processing for 2D FISH so that much of the 3D structure is maintained. Deconvolved sections from z-series through the nucleus are shown.
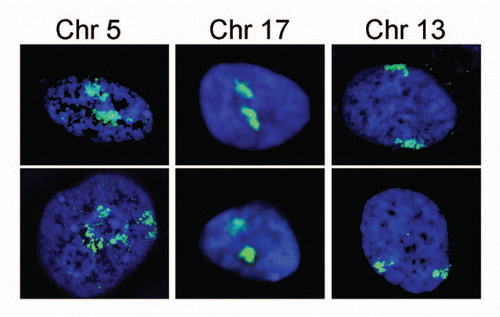
Figure 3 Speculative affinity mechanism for establishment of spatial chromosome organizational patterns. (A) During mitosis the NE either breaks down into ER /NE vesicles or diffuses into the ER. The vesicle model is shown here. Distinct vesicles contain specific components (red and green triangles representing different NETs). Components of some vesicles interact with regions of particular condensed chromosomes. (B) At the end of mitosis the NE starts to reform from vesicles with some specific chromosomes still being attached to particular vesicles. (C) The NE has reformed with some chromosomes being trapped at the NE due to specific NE components that have a high affinity for these chromosomes. The chromosomes are still largely condensed. (D) During interphase the chromosomes decondense and occupy distinct territories within the interphase nucleus. At this point a multitude of less specific lower affinity interactions from the lamina would be expected to help maintain the positioning established in mitosis.
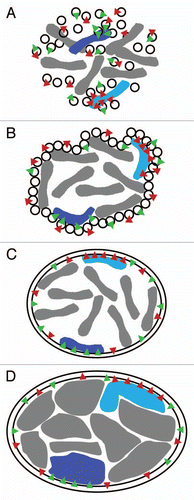
Figure 4 Elastic behavior of the NE. If the intermediate filament lamin polymer supporting the NE were very stiff like other filament systems in the cytoplasm, it would be likely to rupture under the stresses exerted on it by the genome (e.g., growth during replication or rapid movements of regions within chromosome territories (A–C) or the cytoskeleton (D–F). When all components are bound and working together with an elastic nucleoskeleton the whole system can move slightly together while providing a counterforce to that exerted (A and D). In contrast, if the nucleoskeleton functioned like a brick and mortar scaffolding, then components of the system might pull apart from one another or the lamin polymer might physically rupture as do microtubules when subjected to strong bending forces (B, C, E and F). In this case NETs that have strong interactions with chromatin and/or the lamin polymer might even be pulled out of the membrane.
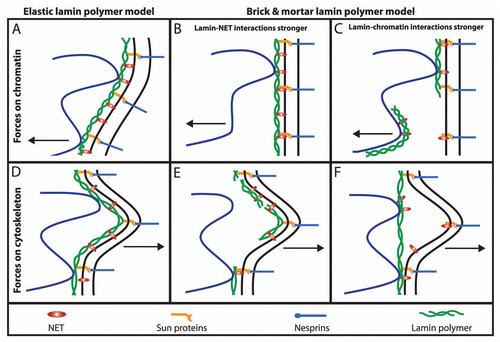
Figure 5 Transcription factories in the nuclear interior can be affected by NE affinity for distinct regions on chromosomes. Because each chromosome is essentially a continuous strand of DNA folded over on itself for compaction into the 10 or 30 nm fibers observed by electron microscopy, it can be unraveled or compacted based on connections to the NE. (A) Recent 3C and 4C chromatin capture studies have revealed interactions within chromosomes and between adjacent chromosomes (e.g., chromosomes A–C in the diagram). Some of these are thought to act as transcription factories where greater local concentrations of transcriptional proteins can optimize transcription (larger green arrows). The availability of chromosome regions to participate in such transcription factories may depend on connections with chromatin and the INM (regions marked i and ii). (B) Changing the pattern of the connections to the INM (e.g., by recruiting also locus iii to the NE) will also affect the transcription factory structure and transcription levels.
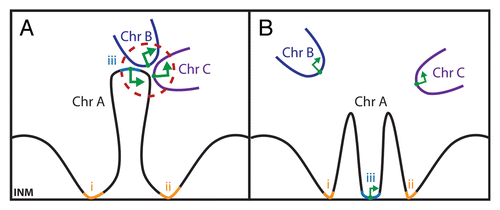
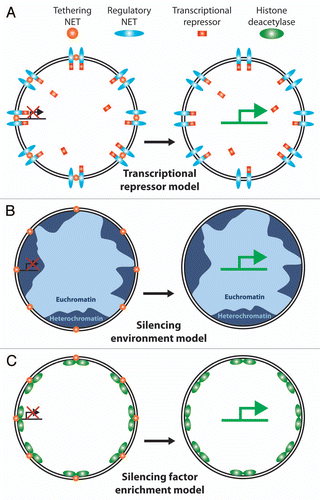
Figure 6 Possible mechanisms for NE interactions to regulate gene expression. Various models have been proposed for how NE tethering of a chromosome or gene could affect gene regulation. (A) Transcriptional regulator sequestration. One NET (orange ball) recruits a gene to the periphery while another NET (blue oval) recruits a transcriptional regulator, in this case a transcriptional repressor (red rectangle). Because the environment of the NE is only ∼1/40 the volume of the nucleus this would effectively increase the local concentration of the transcriptional regulator to keep the gene more tightly repressed. During differentiation expression would shut down for the NET tethering the gene (or the transcriptional repressor) enabling the gene to move away from the high local concentration of the repressor and become more strongly activated. There are obviously many versions of this model. (B) Recruitment to a generally silencing environment. The majority of NE interactions with chromatin identified to date appear to involve heterochromatin by both the original definition of electron dense chromatin observed by electron microscopy and the more modern definition of histones carrying silencing modifications. Thus recruitment of a gene to the periphery could result in its silencing by the general environment. One flaw with this model is that NETs tend to be generally distributed throughout the INM and there are also patches of euchromatin at the periphery so the gene could conceivably move to an active region and not be repressed e.g., tethering NETs (gold balls) in lighter blue regions of euchromatin. (C) Silencing enzyme recruitment model. In addition to binding directly to silenced chromatin, some NETs have been found to recruit factors that modify chromatin to a silent configuration (e.g., the LAP2 interaction with HDAC3 and the LBR interaction with MeCP2). Thus merging aspects of the first two models, co-recruitment of a gene and a chromatin-silencing enzyme to the periphery would effectively shut expression from the gene.