Figures & data
Table 1. Nuclear pore proteins in physical interaction with genomic loci
Figure 1. Regulation of glucose-repressed gene expression in Saccharomyces cerevisiae. Regulation of SUC2 expression is well characterized, and the proteins that carry out this regulation have been studied extensively. The SUC2 system therefore provides a good model for understanding how other genes controlled by the same factors are regulated. (A) A kinase-transcription factor pair work together to control expression of SUC2. Left panel. In the presence of glucose (repressed), the C2H2 zinc finger protein Mig1 binds DNA and inhibits transcription of SUC2, plus approximately 300 other genes. Complete inhibition also requires the hexokinase Hxk2. Snf1 kinase, a structural and functional homolog of mammalian AMPK, is found in the cytoplasm. Right panel. When glucose is depleted or withdrawn (derepressed), Snf1 is phosphorylated by upstream kinases, enters the nucleus and phosphorylates Mig1, which is then exported together with Hxk2. For detailed review, see reference Citation48. (B) Chromatin structure of the SUC2 promoter. Left panel. In the presence of glucose (repressed), four nucleosomes (numbered 1 through 4) cover the SUC2 promoter; a gradient of histone H3 acetylation decreases from 5′ to 3′.Citation55 Two GC-rich sequences are required for repression of transcription (red t-bars) from the TSS (dashed black arrow).Citation88 These sites are interchangeably bound in vivo by Mig1 and its homolog Mig2; function of the former, but not the latter, is regulated by the Snf1 kinase.Citation48 The average location of the first Mig1/2 site (left-most red rectangle) is between nucleosomes 1 and 2. The second Mig1/2 site (right-most red rectangle) is close to the end of the DNA covered by nucleosome 2Citation89-Citation91; nucleosome and repressor may compete for occupancy of this site. The TATA box (green rectangle) is covered by nucleosome 4.Citation89-Citation93 Right panel. Full induction of transcription requires the Swi/Snf chromatin remodeling complex.Citation57,Citation90,Citation91,Citation94-Citation98 Swi/Snf associates more stably with promoter nucleosomes that have been acetylated by SAGA and NuA4 complexesCitation57; in the absence of glucose, acetylation of H3 and H4 tails increases for all nucleosomes. The DNA formerly covered by two nucleosomes (nucleosomes 2 and 3) is more frequently covered by only one. Nucleosome 4, covering the TATA box, is hyperacetlyated and becomes unstable.Citation55 Initiation occurs at the TSS (black arrow). Additionally required for full induction of SUC2 transcription are the nucleosome remodeling protein Spt6Citation99,Citation100 and the transcriptional activator Gcr1 (blue oval, Gcr1 binding site).Citation90,Citation101-Citation104 Nuclear pore proteins also interact with the SUC2 promoterCitation13; it is important to learn how they collaborate with the well-characterized transcription factors described here.
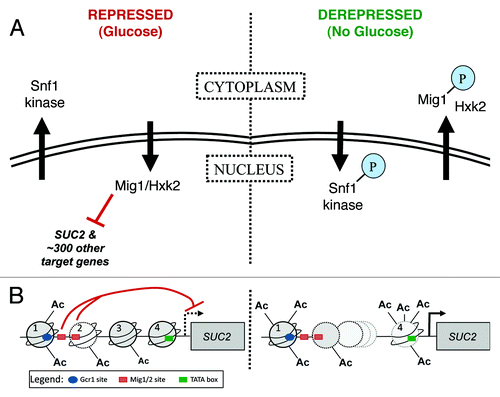
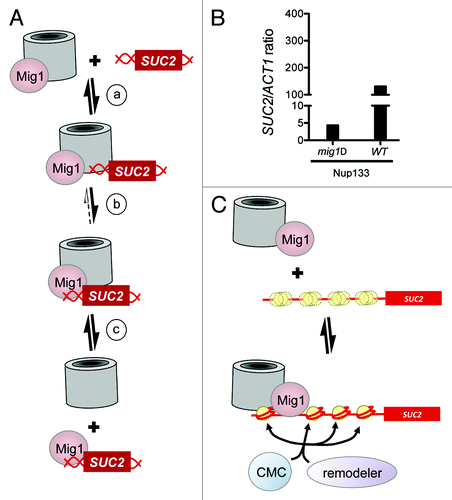
Figure 2. How nucleoporins might influence access to a specific site in DNA. (A) Model for NPC facilitated Mig1 binding to DNA. (a) In the presence of glucose, Mig1 associates with NPCs; the SUC2 locus contacts NPCs transiently. (b) Increased local concentration of both Mig1 and its consensus site facilitates DNA binding by the repressor. (c) The repressor-bound promoter can dissociate from the NPC. (B) Interaction of Nup133 with the SUC2 promoter is reduced in mig1Δ cells. TAP-tagged Nup133 was immunoprecipitated from mig1Δ or wild type cells grown in media containing glucose as the carbon source, then fixed with formaldehyde as described previously.Citation13 Crosslinks were reversed and PCR was used to amplify the promoters of SUC2 and ACT1 (negative control) from recovered material. Amplified target is expressed as a ratio of SUC2/ACT1, normalized to the amount of product amplified whole cell extracts (input). Adding increasing amounts of input DNA shows that amplification of the product is linear. A representative experiment is shown; similar results were obtained for Nup145C (not shown). (C) A model for collaborative nucleosome positioning. Top panel. Mig1 associates with nuclear pores as in (A) above. SUC2 is not associated with nuclear pores, and the nucleosomes across its promoter are poorly positioned (three yellow circles, one distinct and two indistinct, represent the average position of one nucleosome, over time and across a population). Bottom panel. One or more subunits of the nuclear pore associate with the SUC2 locus, serving as a marker or barrier against which chromatin remodelers can position a single nucleosome, thus seeding an ordered array. Alternatively, nuclear pore proteins may direct the activity of chromatin modifiers such as SAGA and NuA4, which in turn direct the activity of remodelers such as Swi/Snf. Models shown in A and C are not mutually exclusive; for example, nuclear pores contribute to nucleosome positioning at step (b) of model A.