Figures & data
Figure 1. Localization of TRAP150 and Btf is dependent on RNA Polymerase II activity. (A) In the absence of α-amanitin, TRAP150 (e, arrow) localizes in nucleoplasmic foci, shows some overlap with splicing factor U1–70K (f) in nuclear speckles and localizes to enlarged rounded speckles (a, arrow) after α-amanitin treatment similar to U1–70K (b, arrow). (B) Btf (m, arrow) localizes in nucleoplasmic foci, shows some overlap with splicing factor U1–70K (n) and localizes to enlarged rounded nuclear speckles (i, arrow) after α-amanitin treatment similar to U1–70K (j, arrow). DNA was stained with DAPI. Boxed regions in a, e, i and m are enlarged in insets. Scale bar, 5 µm.
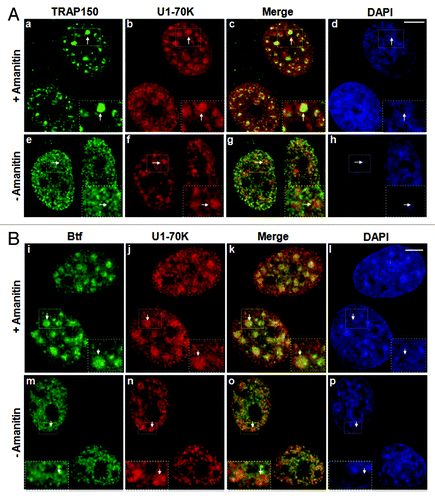
Figure 2. Btf and TRAP150 localize at transcription sites in situ. (A) Schematic representation of the rat β-tropomyosin (BTM) minigene that includes genomic sequence from exon 5 through exon 8. Introns are represented as thin lines connecting the exons. (B) HeLa cells stably expressing the rat BTM minigene were processed for RNA-FISH localization of BTM reporter RNA (b, g) and immunofluorescence localization of SRSF1 (c, h) and either TRAP150 (a) or Btf (f). (C) Schematic representation of U2OS 2–6-3 inducible reporter array.Citation17(D) U2OS 2–6-3 cells transfected with YFP-LacR and pTetON were left uninduced (o-r, w-z) or induced to activate the reporter (+Dox; k-n, s-v). At 2.5 h after dox addition, the cells were processed for immunofluorescence localization of TRAP150 (k-r) or Btf (s-z). DNA was stained with DAPI. Boxed regions in k and o are enlarged in insets. Scale bar, 5 µm.
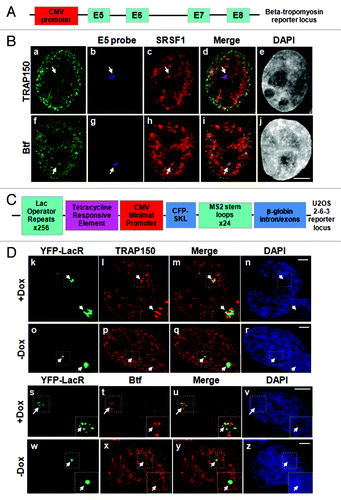
Figure 3. Localization of Btf and TRAP150 at transcription sites is dependent on RNA polymerase II activity. (A) In the absence of amanitin, TRAP150 (c) localized at the active locus. In the presence of amanitin, the locus was decondensed (e); however, MS2BP accumulation in speckles rather than at the active locus indicated that no transcripts were produced under these conditions (f) and TRAP150 (g) was absent. (B) In the absence of amanitin, Btf (j) localized at the active locus. In the presence of amanitin, the locus was decondensed (m); however, MS2BP accumulation in speckles rather than at the active locus indicated that no transcripts were produced under these conditions (n) and Btf (o) was absent. Boxed regions in a, e, i and m are enlarged in insets. (C) Btf and TRAP150 do not colocalize with transcriptional activator at the U2OS 2–6-3 reporter locus. U2OS 2–6-3 cells were transiently transfected with pTetON-YFP and CFP-LacR and seeded in the presence of doxycycline. After 2.5 h, cells were processed for immunolabeling of endogenous Btf or TRAP150. 100% of the cells counted showed that Btf (a) or TRAP150 (e) accumulate at the active locus (panel b and f) but does not colocalize with the transcriptional activator encoded by pTetON (rtTA; c and g). Boxed regions in b and f are enlarged in insets. Scale bar, 5 µm.
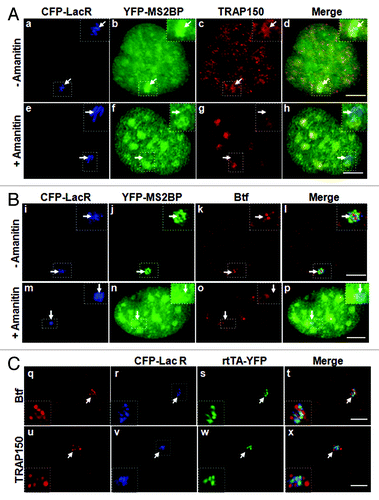
Figure 4. Btf and TRAP150 colocalize with each other, with splicing factor SRSF1, and most precisely with Magoh at transcription sites in situ. (A) U2OS 2–6-3 cells expressing pTetON, YFP-MS2BP (panel a) and CFP-TRAP150 (panel b) were processed for immunolabeling of Btf (panel c). 50 out of the 50 loci scored showed that Btf and TRAP150 overlap extensively at the active locus. (B) U2OS 2–6-3 cells expressing YFP-LacR, pTetON and either CFP-Btf or CFP-TRAP150 were fixed 2.5 h after doxycycline addition and processed for immunolabeling of SRSF1 (a-l). 50 out of the 50 loci scored showed colocalization of Btf and TRAP150 with SRSF1 at the active locus. (C) U2OS 2–6-3 cells stably expressing mcherry-LacI (m, q) were transfected with YFP-Magoh (o, s) and either CFP-Btf (n) or CFP-TRAP150 (r). 50 out of the 50 loci scored showed remarkable colocalization of Btf and TRAP150 with Magoh at the active locus. Boxed regions in a, e, i, m and q are enlarged in insets. To avoid saturating image pixel intensity for CFP-Btf/CFP-TRAP150 at the region of the locus, low image exposure times were required; therefore, the nuclear CFP-Btf/CFP-TRAP150 that was also present in nucleoplasmic foci does not appear in these images. Scale bar, 5 µm.
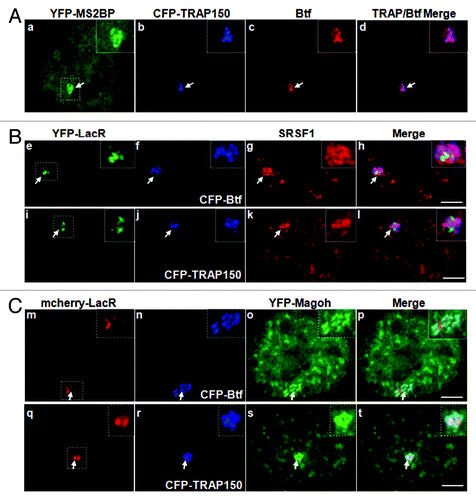
Figure 5. Btf depletion increases levels of BTM transcripts in the cytoplasm. (A) Immunoblots from cells treated in parallel confirm depletion of Btf or TRAP150. Beta-actin was used as a loading control. (B) Validation of nuclear/cytoplasmic RNA fractionation by RT-PCR using primers targeting human U2 snRNA (nuclear) and S14 (cytoplasmic) on total (lane T), nuclear (lane N) and cytoplasmic (lane C) RNA samples. (C) Representative qRT-PCR analysis shows the relative change in the BTM transcripts in nuclear vs. cytoplasmic compartments when normalized to GAPDH. Grey bars represent nuclear BTM RNA while black bars represent cytoplasmic BTM RNA. *p values ≤ 0.05; #p values ≤ 0.1.
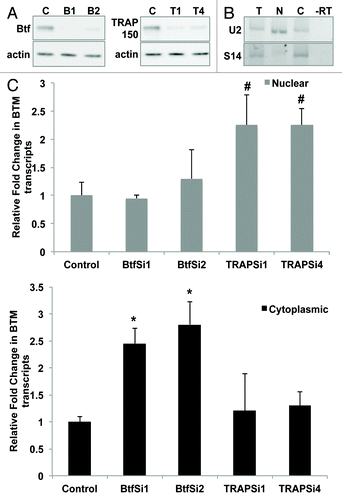
Figure 7. Cytoplasmic polyadenylated RNA increases following depletion of Btf but not TRAP150. HeLa cells were transfected with control siRNA, (A) siRNA oligos specific for Btf (BtfSi1 and BtfSi2) or (B) siRNA oligos specific for TRAP150 (TRAPSi1 and TRAPSi4). Cells were processed 72 h post-transfection for RNA-FISH using Texas Red-conjugated oligo-dT50 to visualize polyadenylated RNA. (C) Immunoblots and corresponding densitometry plots showing relative expression levels of Btf or TRAP150. Beta-actin was used as a loading control. Data set shown is a representative of three independent experiments having similar results. Scale bar, 5 µm.
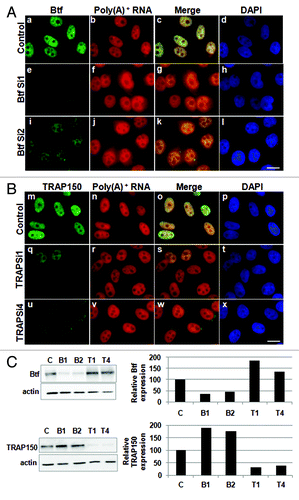
Figure 8. Cytoplasmic polyadenylated RNA increases in Btf-depleted cells when compensatory upregulation of TRAP150 is blocked. (A) HeLa cells were transfected with control siRNA or a mixture of BtfSi1 and TRAP150Si4. 72 h post transfection, the cells were processed for RNA-FISH using Texas Red-conjugated oligo-dT50 to visualize polyadenylated RNA distribution. (B, C) Immunoblots and corresponding densitometry plots showing relative expression levels of Btf or TRAP150. Beta-actin was used as a loading control. Densitometry shows the average of two replicate immunoblots. (D) Graph of nuclear vs. cytoplasmic ratio of polyadenylated RNA. Total fluorescence measurements were performed using ImageJ. Each bar represents the average ratio of pixel intensity for nuclear poly(A)+ RNA to pixel intensity for cytoplasmic poly(A)+ RNA in 25 cells from 3 independent experiments. Error bars represent s.d. of fluorescence intensities. *p values ≤ 0.05. Scale bar, 5 µm.
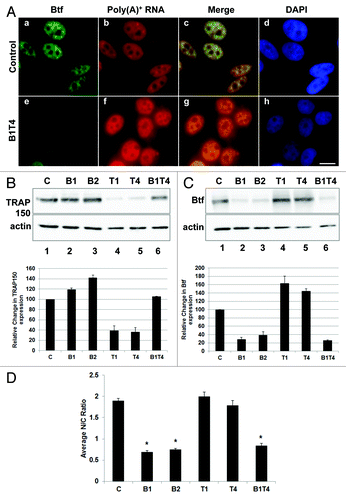
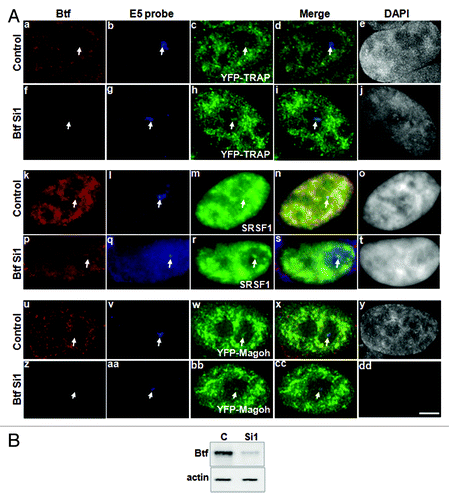
Figure 6. Depletion of Btf does not prevent recruitment of TRAP150 or Magoh to the BTM transcription site. (A) BTM HeLa cells were transiently transfected with YFP-TRAP150 or YFP-Magoh. Twenty-four hours post-transfection, the cells were again transfected with 60 pmoles of either control siRNA (panels a, k and u) or Btf siRNA 1 (panels f, p and z). 72 h after knockdown, the cells were processed for RNA-FISH localization of reporter RNA using oligos against exon 5 (panels b, g, l, q, v and aa) as describedCitation16 and immunofluorescence localization of Btf, splicing factor SF2/ASF or GFP. Arrows indicate the BTM transcription site. DNA was stained with DAPI. Scale Bar, 5 µm. (B) Immunoblot confirming depletion of Btf or TRAP150. Actin was used as a loading control.