Figures & data
Figure 1.S. pombe nucleoporins fused to GFP. (A) Subcellular localization of GFP-tagged nucleoporins. The different images were obtained with the same acquisition times and processed in parallel. Deconvolved images are shown. The top left two panels show fluorescence (left) and bright field (right) images of an spCut11-GFP expressing cell. Nucleoporins were classified into seven groups according to localization in the NPC inferred from localization of the budding yeast orthologs. The scale bar represents 10 μm. (B-D) Localization of GFP-spSec13 (B), spAmo1-GFP (C), and spTts1-GFP (D) in wild type and cells lacking spNup132 (nup132Δ); in (B), GFP was fused with N-terminus (left) and C-terminus (right) of spSec13. spCut11-mCherry was simultaneously observed as a known nucleoporin marker. Yellow arrows indicate NPC-clustering regions in nup132Δ cells (lower panels). The scale bars represent 10 μm.
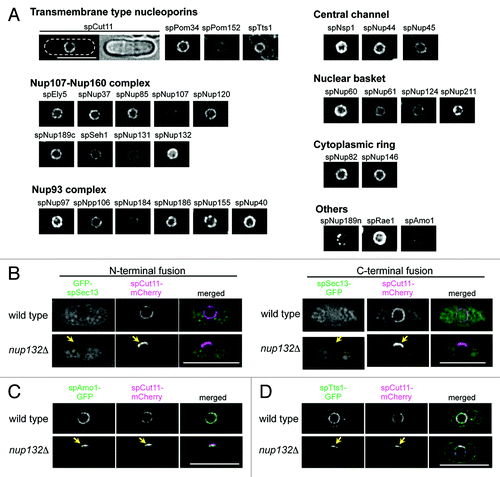
Figure 2. Quantification of GFP-tagged nucleoporins. (A) Fluorescence intensities of GFP-tagged nucleoporins. Fluorescence intensities of about 50 cells from each strain cultured at 26 °C were measured. Average values after background subtraction are shown in the bar graph. Error bars represent standard deviations. Components of the Nup107-Nup160 subcomplex are shown in magenta. (B) Quantitative western blot analysis of Nup107-Nup160 subcomplex nucleoporins. GFP-fused nucleoporins were fractioned by SDS-PAGE and detected by anti-GFP antibody (left). The membranes were stripped and reprobed to detect endogenous spNup189n as an internal control (right). The numbers above the images indicate cell numbers used to prepare the whole cell extract for each lane. (C) The relative amounts of Nup107-Nup160 subcomplex nucleoporins based on quantitative western blot analysis. Protein band intensities for 1 × 106 cells in (B) were measured for quantification. See Materials and Methods for detail.
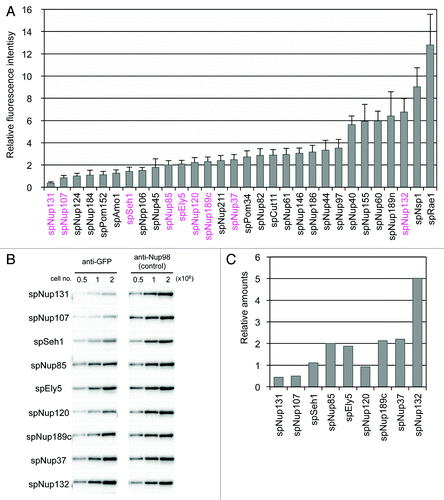
Table 1. Nucleoporins in S. pombe
Table 2. Essentiality of S. pombe nucleoporins
Figure 3. Fluorescence intensities of GFP-tagged nucleoporins measured by HILO microscopy. (A) HILO microscopy images of cells expressing spNup131-GFP, spNup107-GFP, spNup85-GFP, and spNup132-GFP. The nuclear region of a single cell is shown. The scale bar indicates 1 μm. (B) Fluorescence intensity of nucleoporins in single NPCs. Average and standard deviation are shown.
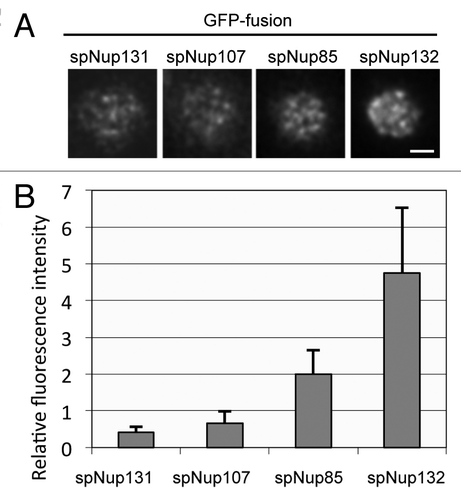
Table 3. Correspondence table of nucleoporins
Table 4. Function of non-essential nucleoporins for various growth conditions
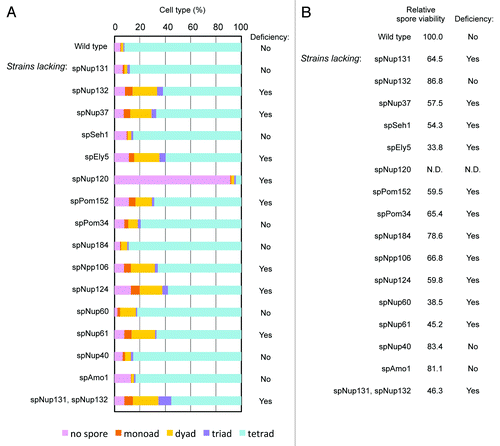
Figure 4. Effect of gene disruption of non-essential nucleoporins on spore formation. (A) Spore numbers in wild type and nucleoporin mutants. Cells were cultured on sporulation medium for two days, and zygotic cells containing spores were differentially scored. “Yes” and “No” indicate cell strains with a deficiency and no deficiency in spore formation, respectively. (B) The numbers indicate the spore viability of wild type and nucleoporin mutants. Cells forming spores were treated with β-glucuronidase to digest vegetative cells, and the resistant spores were counted and spread on growth media. After colony formation, colony numbers were counted and spore viability was calculated. The value was normalized with that of the wild type, which was given a value of 100. “Yes” and “No” indicate cell strains with a deficiency and no deficiency in spore viability, respectively.