Figures & data
Figure 1. Epigenetic control of adaptive immune cell differentiation and function. Adaptive immune system consists in the cellular (T cell mediated) and humoral (B cell mediated) arm. Differentiation of adaptive immune cells occurs initially in primary lymphoid organs (thymus for T cells and bone marrow, BM, for B cells), where each progenitor cell (Pre-T and Pre-B) rearranges its antigen receptor loci at the DNA level in order to express a different receptor. In these organs only cells expressing a functional receptor not recognizing self-antigens receive anti-apoptotic stimuli (positive selection) and survive from negative selection (depletion of autoreactive clones). Upon rearrangement and differentiation effector and regulatory mature cells (matT helper, matTh, or matT cytoxic, matTc, if expressing CD4 or CD8 as TCR co-receptor, respectively, matB and thymic derived CD4+ T regulatory, tTreg) migrate to secondary lymphoid organs (mainly spleen and lymphnodes), where naïve (nTh, nTc, nB, and tTreg) cells can encounter an antigen (Ag). Ag-TCR or Ag-BCR interaction induces pattern of expressions leading to the generation of activated (and thus functional) immune cells. TCR recognizes Ag only if processed and presented by antigen presenting cells (APC) through Major Histocompatibility Class (MHC) molecules, while BCR recognizes free Ag. Depending on the type of Ag and/or the MHC presenting it, naïve cells differentiate in different subsets with different functions: (1) nTh differentiate mainly in Th1, Th2, Th17 cells that produce different cytokines inducing different B cell- mediated or myeloid response to extracellular pathogens; (2) in case of non-harmful Ag, nTh differentiate into T regulatory cells (peripheral, pTreg) that, together with tTreg cells, induce tolerance and dampen immune response through different mechanisms; (3) nTc differentiate into Cytotoxic T Lymphocyte (CTL) that mediate killing of the infected target cells; (4) nB cells differentiate into activated B (actB) cells and this process requires antibody maturation and differentiation through class switch recombination (CSR) and somatic hyper mutation (SHM), respectively; actB cells further differentiate in plasma cells (PC) that are factories for the secretion of antibodies (Ab). Most of the expanded activated cells undergo apoptosis after pathogen clearance while a small fraction persists in the body as memory cells (mTh, mTc, and mB) and mediate faster secondary response in case of re-encountering of the Ag. The adaptive immune cell processes controlled at the epigenetic level are highlighted in stars with relative references.
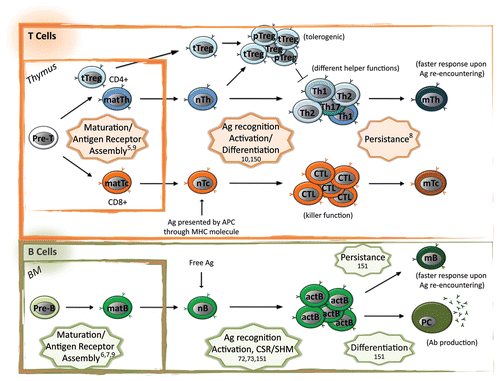
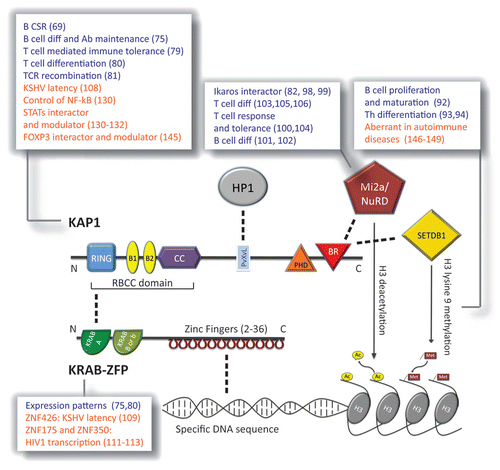
Figure 2. Function(s) of the KRAB proteins. Schematic of the main domains of KAP1 (top) and a prototypic KRAB-ZFP (bottom), and of the main molecules interacting with each of the two (direct interaction is depicted as dashed line). KAP1 domains (N to C-terminal). RING, Really Interesting New Gene zinc finger; B1 and B2, B box zinc fingers; CC, coiled coil. RING, B boxes, and CC form together the RBCC domain, needed by KAP1 to interact with the KRAB A domain of the KRAB-ZFP. PvXvL, hydrophobic pentapeptide needed for interaction with HP1 (Heterochromatin Protein 1); PHD, plant homeo-domain; BR, bromodomain. PHD and BR cooperate in binding the Mi2a/NuRD deacetylase complex and the SETDB1 histone 3 lysine 9 methyl transferase. Once recruited by KAP1 these two histone-modifying factors are able to induce heterochromatin formation by modifying histone 3 (H3) tail. KRAB-ZFP domains (N to C-terminal). KRAB A, Krüppel-associated box A. It is a transcriptional repressor module present in all KRAB-ZFP and mediates KAP1 recruitment. The second KRAB box is facultative and can be B or b depending on the primary structure of the sequence. Zinc fingers are present in tandem repeats and can vary from 2 to 36 in the family; they are able to bind specific DNA sequences. KAP1 has not a DNA-binding domain; KRAB-ZFP is thus the linkage between specific DNA stretches and KAP1-mediated complexes. Listed in the pop-ups the roles of the depicted protein or histone modification as assessed in the mouse (in blue) and in the human (in orange) adaptive immune system; in brackets the relative references.