Figures & data
Figure 1. Experimental set up. At day 0, T-cell receptor (TCR)-transgenic SCID mice were injected s.c. with MOPC315 myeloma cells (green cells) suspended in liquid Matrigel. When the Matrigel solution reached body temperature, it gelified and formed a plug embedding myeloma cells. Tumor-specific CD4+ T cells (red cells) became activated in the tumor-draining lymph node (LN), differentiated into TH1 cells, and subsequently migrated to incipient tumor sites (Matrigel plug). Eight d after the injection of tumor cells, mice were euthanized, and tumor-draining LNs and Matrigel plug were dissected out. The in vivo activation of tumor-specific CD4+ T cells was characterized by flow cytometry (in draining LNs and incipient tumor sites) and gene expression profiling (in draining LNs only).
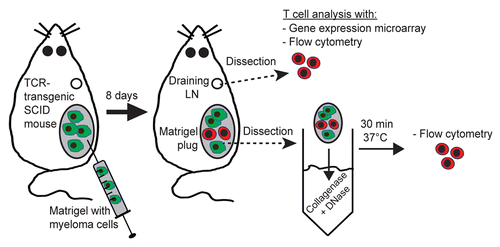
Figure 2. Expression pattern of molecules on the surface of tumor-specific CD4+ T cells in draining LN after in vivo activation. (A–C) T-cell receptor (TCR)-transgenic SCID mice (n = 6–12) were injected s.c. with MOPC315 myeloma cells. Eight d later, the activation of tumor-specific (GB113+) CD4+ T cells from pooled tumor-draining lymph nodes (LNs) was analyzed by flow cytometry (blue curves). Filled gray areas indicate isotype-matched control stainings of activated T cells. For comparison, naïve tumor-specific CD4+ T cells from pooled LNs from non-injected TCR-transgenic SCID mice are shown (black curves). (A) Surface molecules that were upregulated after activation. (B) Surface molecules that were downregulated after activation. (C) Surface molecules that were expressed at similar levels on naïve and activated tumor-specific CD4+ T cells. Data are representative of 2–4 experiments.
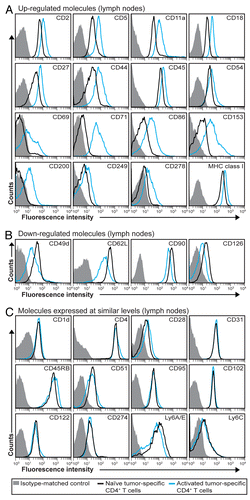
Figure 3. Molecular changes on the surface of activated tumor-specific CD4+ T cells that have migrated to incipient tumor sites. (A and B) T-cell receptor (TCR)-transgenic SCID mice (n = 6–12) were injected s.c. with MOPC315 myeloma cells in Matrigel. Eight d later, the activation of Matrigel-infiltrating, tumor-specific (GB113+) T cells was analyzed by flow cytometry (red curves). Filled gray areas indicate isotype-matched control stainings of Matrigel-infiltrating T cells. For comparison, activated (blue curves) and naïve (black curves) tumor-specific CD4+ T cells from pooled lymph nodes (LNs) are shown. (A) Surface molecules that were upregulated after activation. (B) Surface molecules that were downregulated after activation. Data are representative of 2–4 experiments.
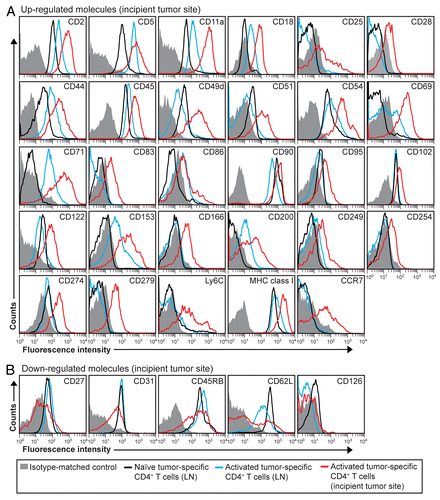
Figure 4. Intracellular cytokine expression in activated tumor-specific CD4+ T cells. (A and B) T-cell receptor (TCR)-transgenic SCID mice (n = 6–12) were injected s.c. with MOPC315 myeloma cells in Matrigel. Eight d later, cytokine expression in tumor-specific (GB113+) CD4+ T cells was analyzed by intracellular flow cytometry, upon stimulation in vitro with ionomycin/phorbol myristate acetate (PMA) in the presence of monensin for 4 h. (A) Cytokine expression in activated tumor-specific CD4+ T cells in tumor-draining lymph nodes (LNs) (blue curves). For comparison, naïve tumor-specific CD4+ T cells from pooled LNs are shown (black curves). (B) Cytokine expression in tumor-specific (GB113+) T cells at incipient neoplastic lesions (Matrigel plugs) (red curves). Filled gray areas indicate isotype-matched control stainings. Data are representative of 2–3 experiments.
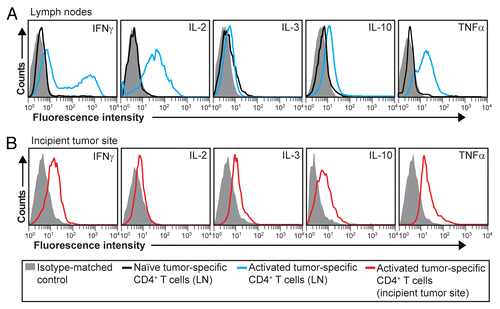
Figure 5. Early activation markers for tumor-specific CD4+ T cells. T-cell receptor (TCR)-transgenic SCID mice (n = 6–12) were injected s.c. with MOPC315 myeloma cells. Six d later, the activation of tumor-specific (GB113+) CD4+ T cells from pooled tumor-draining lymph nodes (LNs) was analyzed by flow cytometry (blue curves). Filled gray areas indicate isotype-matched control stainings of activated T cells. For comparison, naïve tumor-specific CD4+ T cells from pooled LNs from non-injected TCR-transgenic SCID mice are shown (black curves). Data are representative of 2 experiments.
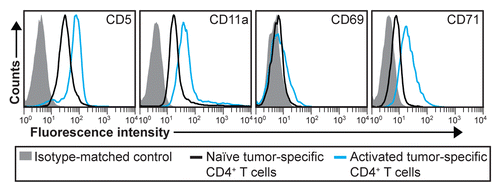
Figure 6. Gene expression data arranged in functional pathways. (A and B) T-cell receptor (TCR)-transgenic SCID mice were injected s.c. with MOPC315 cells. Gene expression in activated tumor-specific CD4+ T cells isolated from pooled tumor-draining lymph nodes (LNs) 8 d later was compared with that of naïve tumor-specific CD4+ T cells. Genes were manually annotated into functional pathways. Genes important for or potentially associated with immune responses are raised from the pie chart. (A) Genes that are upregulated in activated tumor-specific CD4+ T cells as compared with naïve tumor-specific T cells. (B) Genes that are downregulated in activated tumor-specific CD4+ T cells as compared with naïve tumor-specific T cells.
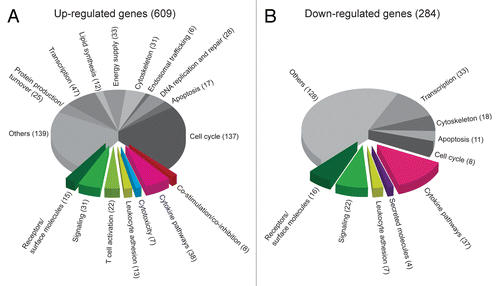
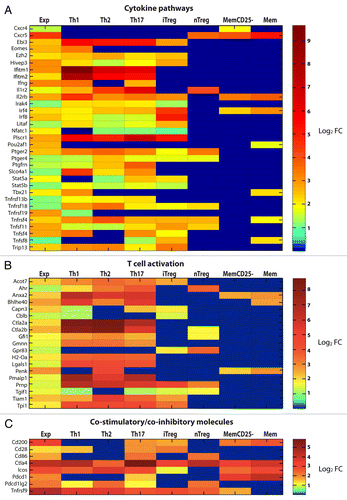
Figure 7. Comparison of upregulated genes in activated tumor-specific TH1 cells to reference data sets relative to several CD4+ T-cell subsets. (A–C) T-cell receptor (TCR)-transgenic SCID mice were injected s.c. with MOPC315 cells. Gene expression of activated tumor-specific CD4+ T cells isolated from pooled tumor-draining lymph nodes (LNs) 8 d later was analyzed. Upregulated genes were determined by comparing the gene expression in (1) activated tumor-specific CD4+ T cells to naïve tumor-specific CD4+ T cells (first column, annotated “Exp”) and (2) various activated reference CD4+ T-cell subsets to naïve reference CD4+ T cells. These reference sets were mouse CD4+ T cells from a previous study (annotated as TH1, TH2, TH17, iTreg and Treg in the heat-map) and memory T cells from the Immunological Genome Project (annotated as MemCD25-, and Mem in the heat-map). Genes that were upregulated (fold change, FC ≥ 2) in tumor-specific TH1 cells were selected for comparisons to reference data sets. Heat-map comparisons were categorized as the manual functional annotation categories shown in pie charts (). Each column represents expression comparisons for each CD4+ T-cell subset, and the scale bar indicates the level of differential gene expression (log2 transformed). Comparisons associated with a FC < 2 are in blue. (A) Genes involved in cytokine pathways. (B) Genes involved in T-cell activation. (C) Genes involved in co-stimulation/co-inhibition. Additional heat-maps can be found in Figures S1 and S2.