Figures & data
Figure 1 EGFP expression in E15.5 pancreas. (A) Stereomicroscopic view of pancreas/gut dissection from an E15.5 ngn3-EGFP mouse embryo. (B) The same pancreas/gut dissection using EGFP fluorescent filters. D, duodenum; St, stomach; Sp, spleen; Dp, dorsal pancreas; Vp, ventral pancreas.
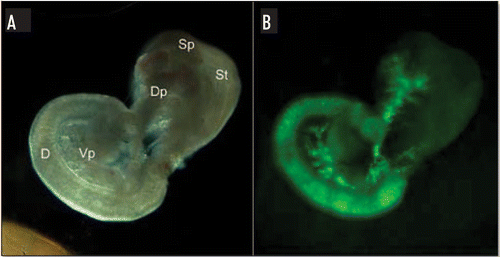
Figure 2 Genotyping the ngn3-EGFP 1.37 line. Genotyping PCR was carried out on tail tip DNA to check for the presence of both EGFP (483 bp) and neomycin (neo) (410 bp) constructs in the offspring of the founder mice. +/+, positive for EGFP and Neomycin transgenes; −/−, negative for EGFP and Neomycin transgenes.
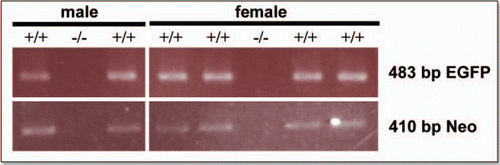
Figure 3 EGFP fluorescence colocalizes with anti-EGFP staining in the E15.5 pancreas. In the panels marked EGFP the fluorescence was detected using EGFP filters. In panels marked anti-EGFP immunohistochemistry was performed on cryosections using an anti-EGFP antibody and a Texas Red conjugated secondary antibody with DAPI staining to identify nuclei. The merged image was obtained by merging images using Photoshop 6.0 software (Scale bar for: upper panels = 100 µm, lower panels = 20 µm).
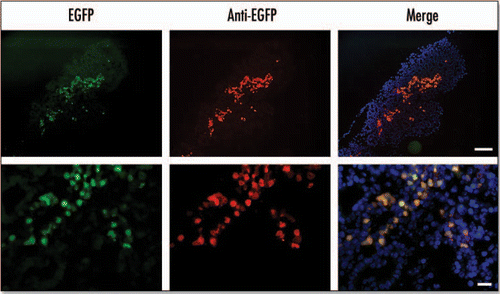
Figure 4 EGFP colocalizes with ngn3 at E15.5. EGFP fluorescence was measured using an EGFP filter. Ngn3 was localized by immunohistochemistry of cryosections using an anti-ngn3 antibody and AlexaFluor 350 conjugated secondary antibody. Arrows indicate cells in which EGFP and ngn3 colocalize. The arrow-head indicates auotofluorescence associated with a blood cell (Scale bar = 50 µm).
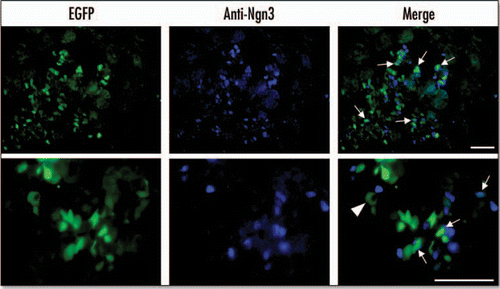
Figure 5 Localisation of EGFP in the E15.5 pancreas to regions enriched in endocrine markers. Immunohistochemistry was performed on cryosections of E15.5 embryonic pancreas dissections. Primary antibodies for insulin (INS) and glucagon (GLU) were visualized with an anti-mouse Rhodamine conjugated secondary antibody. Primary antibodies for amylase were visualized using anti-rabbit AlexaFluor 350 secondary antibody. EGFP was visualized directly using an EGFP filter (Scale bars = 50 µm).
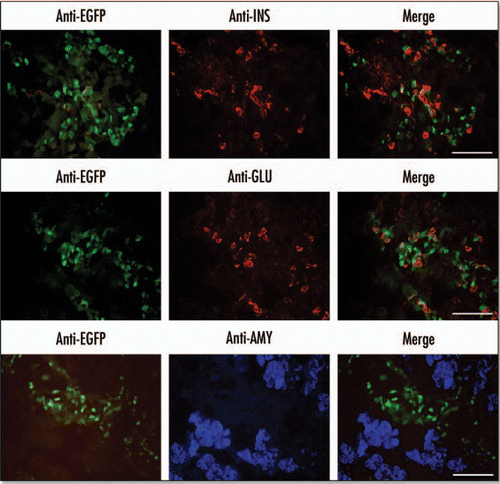
Figure 6 EGFP expression in the E18.5 pancreas. Immunohistochemistry was performed on cryosections from an E18.5 pancreas. Anti-ngn3 primary antibody and AlexaFluor 350 secondary antibody were used to detect Ngn3 expression. EGFP was visualized directly using an EGFP filter (Scale bar = 50 µm).
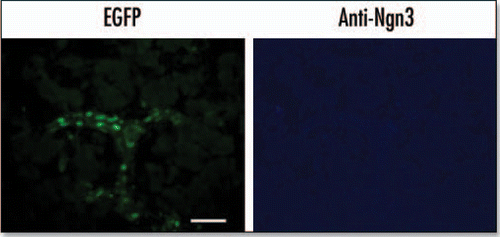
Figure 7 EGFP expression in endocrine-enriched regions of the E18.5 pancreas. Immunohistochemistry was performed on cryosections from an E18.5 pancreas. Primary antibodies for insulin (INS) and glucagon (GLU) were visualized with an anti-mouse rhodamine conjugated secondary antibody. Antibodies against amylase (AMY) were visualized using anti-rabbit AlexaFluor 350 secondary antibody. EGFP was visualized directly using an EGFP filter (Scale bar = 50 µm).
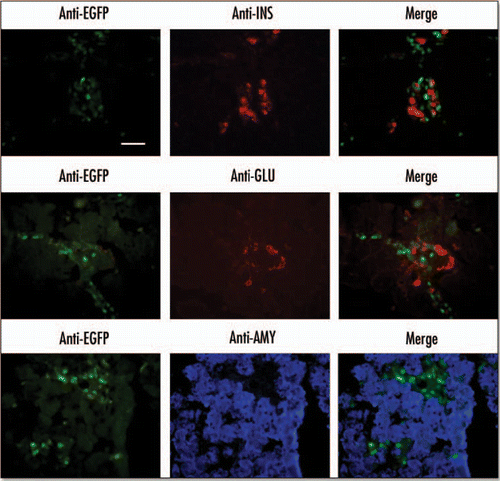
Figure 8 Fluorescence activated cell sorting (FACS) of EGFP positive cells from an E15.5 pancreas. Dissected pancreases from a litter of transgenic mice were dispersed into a single cell suspension via enzyme digest and sorted based on their EGFP fluorescence. Analysis of the cells' scatter (A) shows two distinct and viable populations. The EGFP-positive population (gated green population) can be sorted (B) from the background population, which is shown as a white diagonal line of events. (C) A histogram analysis of the purified EGFP-negative population (gray). (D) A histogram of the purified EGFP-positive population (green).
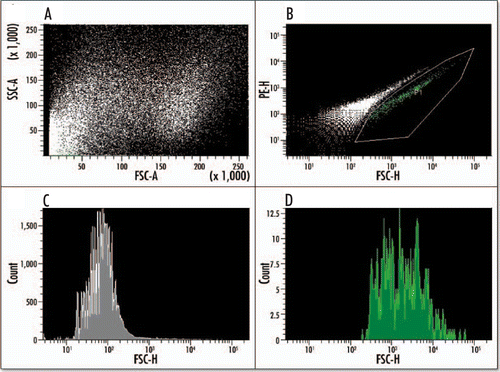
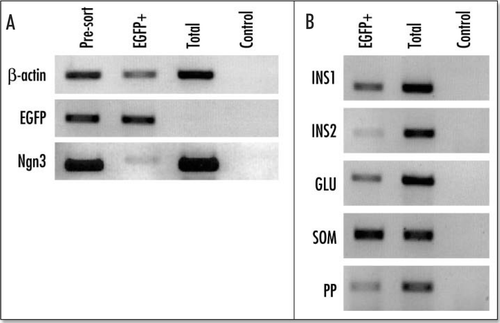
Figure 9 Reverse transcriptase PCR (RT/PCR) analysis of the purified EGFP-positive cells from an E15.5 pancreas. Ethidium bromide stained agarose gel of RT/PCR products from total RNA isolated from presorted FACS purified EGFP-positive cells, and the total population of enzyme dispersed cells from EGFP-negative embryos from the same litter. The control panel shows the reaction in the absence of reverse transcriptase. (A) b actin, EGFP and ngn3; (B) The results for insulin 1 (INS1), insulin 2 (INS2), glucagon (GLU), somatostatin (SOM) and pancreatic polypeptide (PP).