Figures & data
Figure 1. Schematic presentation of stem cells on RGD nanopatterns with a non-fouling background. RGD ligands are grafted onto gold nanodots, which are covalently bound to a PEG hydrogel. Integrins as the receptors of RGD will be linked to the nanodots after cell culture. The pre-designed nanospacing then determines the nanospacing of integrins, and might regulate adhesion, proliferation, and differentiation of MSCs.
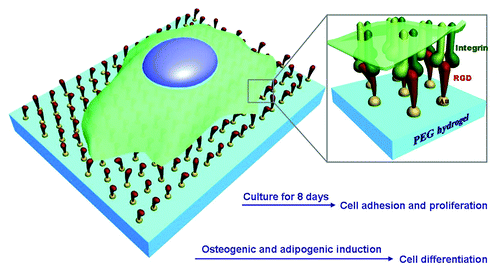
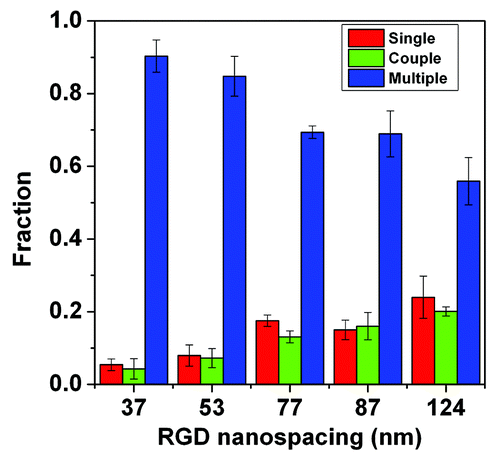
Figure 2. AFM images of gold nanopatterns on glass. The lower row shows the height profiles with respect to the lines in the upper images, indicating that the particle size was around 10 nm. From the left to right, the average nanospacings of gold particles are 34 nm, 48 nm, 70 nm, 79 nm, and 113 nm, respectively. Note: after the nanoarrays were transferred onto the surfaces of PEG hydrogels and then soaked in an aqueous environment to culture cells, the average nanospacings were enlarged to 37 nm, 53 nm, 77 nm, 87 nm, and 124 nm, respectively, considering the swelling ratio of the hydrogels as 1.1.
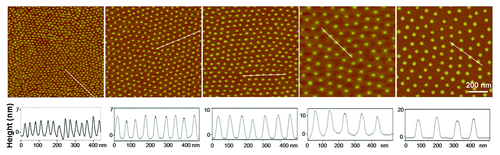
Figure 3. Fluorescence images of stem cells adhering on the RGD patterned PEG hydrogel after 1 d and 8 d of culture. Due to the persistent resistance of the PEG hydrogel to non-specific cell adhesion, cells did not progress past the dip-line during the examination period of 8 d.
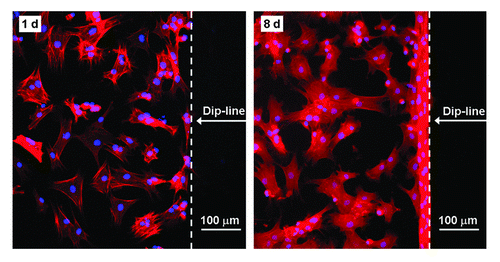
Figure 4. Distributions of cell area, cell circularity, and cell aspect ratio on patterned surfaces of RGD peptides on the PEG hydrogels with indicated nanospacings. The means and standard deviations are marked inside the graphs.
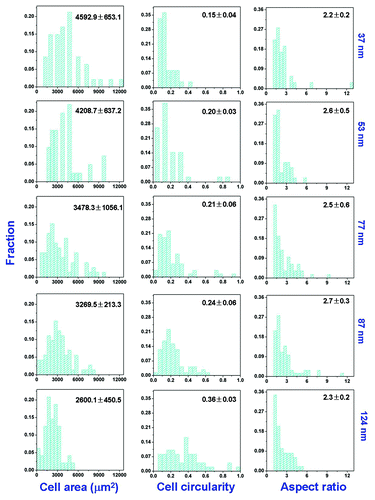
Figure 5. Densities of MSCs adhering on nanopatterned surfaces of indicated RGD nanospacings. Cells were cultured in growth medium for 1 d (GM 1 d) or 8 d (GM 8 d), or cells underwent one-day culture in GM followed by 7 d in osteogenic medium (OM 1+7 d) or adipogenic medium (AM 1 + 7 d). In all cases, the cell-division inhibitor aphidicolin was added at a concentration of 0.5 μg/mL. Insets show proliferation rates calculated by cell density at day 8 over that at day 1. The error bars of the proliferation rates were obtained by an error transfer formula (see Materials and methods).
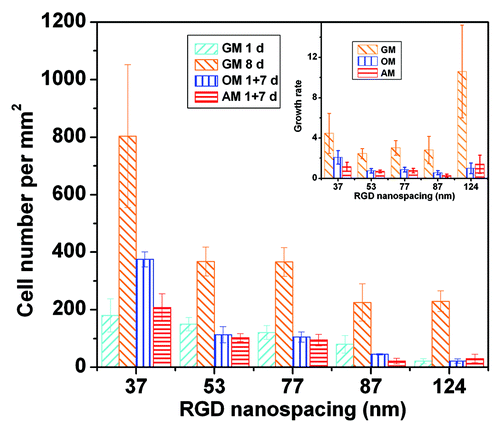
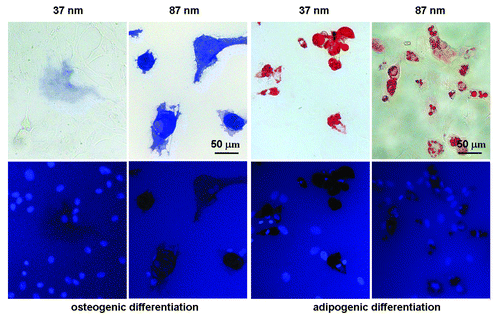
Figure 6. Bright-field micrographs (upper row) and fluorescence micrographs (lower row) of MSCs on RGD nanopatterns of indicated nanospacings in osteogenic or adipogenic medium for 7 d. Before observation, the cells upon osteogenic induction were stained for ALP and nuclei, and those upon adipogenesis for oil microdroplets and nuclei. Both osteogenesis and adipogenesis were enhanced with RGD nanospacing.