Figures & data
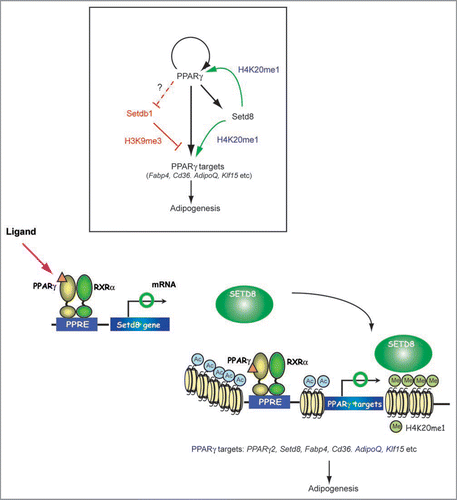
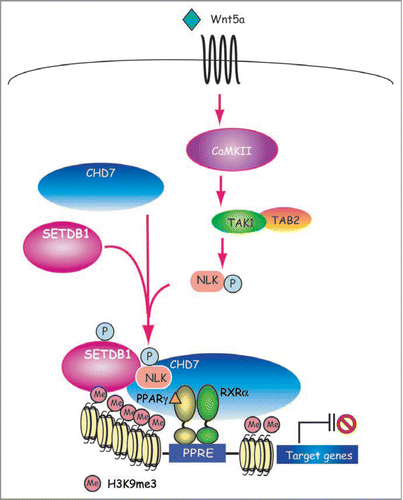
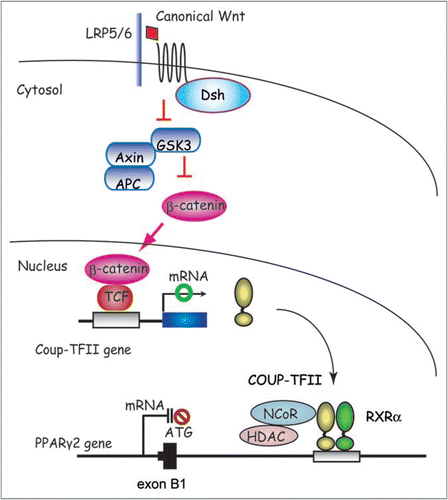
Figure 1 Models for the coordinate regulation of transcription and histone modification by PPARγ for adipogenesis. Two well characterized HKMT, Setdb1 and Setd8 are coordinately regulated by PPARγ and their increased activity facilitates terminal adipocyte differentiation through chromatin modification. PPARγ also drives induction of PPARγ2 via a feedback loop and many other of the target genes via two pathways; one through transcription and the other, by way of an epigenetic pathway. PPARγ requires Setd8 to acquire H4K20me1 modification in order to enhance its transcription, while Setd8 requires PPARγ to be transcriptionally induced. These two are both required for the expression of PPARγ targets. Setdb1 is an anti-adipogenic factor whose expression is downregulated toward the end of differentiation. Setdb1 is also identified as a PPARγ target, however, it remains to be determined whether it is a direct downregulated target genes.