Figures & data
Figure 1 Antiprion drugs as baits in affinity chromatography on immobilized compounds. (A) The structure of the linker connecting the drug to the matrix (Sepharose bead) is crucial. A short spacer can prevent interaction with cellular targets because of steric hindrance (upper scheme). If the spacer is too hydrophobic (depicted in red in the middle panel), it can lead to its auto aggregation thus also preventing target binding. In addition, a hydrophobic linker can generate unspecific hydrophobic interactions with macromolecules from cell lysates. An optimal spacer (lower panel) should be only mildly hydrophobic (hydrophilic part depicted in blue) and long enough to avoid steric hindrance. In this case, specific interactions can occur, as long as the covalent link of the linker to the drug does not affect its antiprion activity too strongly. (B) Possible linker chemical structures are depicted. Polyethylene glycol (PEG) (upper panel) and amino caproylaminopentyloxy linker (lower panel). (C) Affinity chromatography purification on immobilized 6AP. In the example given, the antiprion drug 6AP was covalently liked to Sepharose beads via an amino caproylaminopentyloxy linker. Cell extracts were then incubated with this matrix. After extensive washing, the affinity matrix bound cellular components were analyzed by SDS PAGE followed by silver staining. The interacting proteins were identified by mass spectrometry.
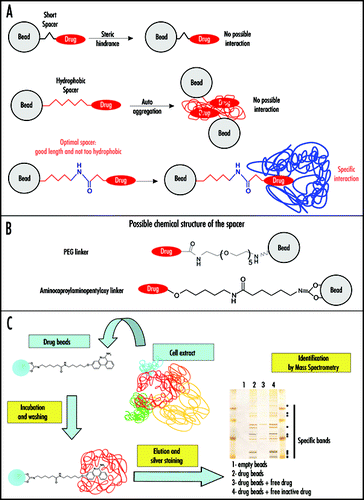
Figure 2 Antiprion drugs as baits in genetic approaches. (A) Genome wide overexpression screen (OES). A cDNA library (of any origin) expressed from the GAL-inducible promoter in a yeast plasmid was transformed in a [PRION+] yeast strain (in the example given [PSI+] forming white colonies) and cells were streaked on a Galactose medium (in condition where strong expression of cDNA was induced) and containing an antiprion drug. The curing effect produces red [psi-] colonies. Colonies which remain white despite presence of the antiprion compound might express a cDNA whose overexpression prevent the curing effect of the drug. These colonies are restreaked on the same Galactose medium containing the antiprion drug to check for resistance to the antiprion drug and also on Glucose medium containing antiprion drug to check that the isolated clone is sensitive to the drug in this condition where expression of the cDNA is largely repressed. In the example given, one clone appears to be a false positive for which resistance is not caused by expression of the cDNA (it remains white on Glucose medium). (B) Three-hybrid assay (Y3H). This method is based on the use of a heterodimeric ligand constituted of a ligand of the receptor used as bait covalently linked to the antiprion drug (6AP in the example given). The heterodimeric ligand can be used to screen against a library of cDNAs (from any origin) in fusion with the activation domain (AD) of a transcriptional activator (here Gal4p). The HIS3 reporter gene will only be activated in cells expressing a cDNA encoding a protein to which the antiprion drug (6AP here) binds, and thus only these cells will be able to grow in a medium lacking histidine.
![Figure 2 Antiprion drugs as baits in genetic approaches. (A) Genome wide overexpression screen (OES). A cDNA library (of any origin) expressed from the GAL-inducible promoter in a yeast plasmid was transformed in a [PRION+] yeast strain (in the example given [PSI+] forming white colonies) and cells were streaked on a Galactose medium (in condition where strong expression of cDNA was induced) and containing an antiprion drug. The curing effect produces red [psi-] colonies. Colonies which remain white despite presence of the antiprion compound might express a cDNA whose overexpression prevent the curing effect of the drug. These colonies are restreaked on the same Galactose medium containing the antiprion drug to check for resistance to the antiprion drug and also on Glucose medium containing antiprion drug to check that the isolated clone is sensitive to the drug in this condition where expression of the cDNA is largely repressed. In the example given, one clone appears to be a false positive for which resistance is not caused by expression of the cDNA (it remains white on Glucose medium). (B) Three-hybrid assay (Y3H). This method is based on the use of a heterodimeric ligand constituted of a ligand of the receptor used as bait covalently linked to the antiprion drug (6AP in the example given). The heterodimeric ligand can be used to screen against a library of cDNAs (from any origin) in fusion with the activation domain (AD) of a transcriptional activator (here Gal4p). The HIS3 reporter gene will only be activated in cells expressing a cDNA encoding a protein to which the antiprion drug (6AP here) binds, and thus only these cells will be able to grow in a medium lacking histidine.](/cms/asset/ab364c31-fac4-4cd9-955f-30206ff2c438/kprn_a_10904053_f0002.gif)