Figures & data
Figure 1 Hsp104 primary structure and location of mutations engineered in this study. The multi-domain structure of the Hsp104 monomer. The N-terminal domain (NTD), nucleotide-binding domains (NBD) 1 and 2, D1 small domain (D1sD), middle region (MR), and C-terminal domain (CTD) are indicated. The domain boundaries are according to Hsp104's alignment with the T. thermophilus ClpB sequence.Citation6 The hsp104 mutations are shown with their allele numbers, for example, A201V is an Ala-to-Val change at position 201.
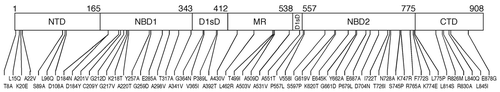
Figure 2 The status of [PSI+] in hsp104 mutants is monitored by color assay of ade1-14 nonsense suppression. Haploid strains carrying the indicated hsp104 mutations and control wild-type [PSI+] strain (AT3) were incubated on YPD at 30°C for four days. The results are summarized in .
![Figure 2 The status of [PSI+] in hsp104 mutants is monitored by color assay of ade1-14 nonsense suppression. Haploid strains carrying the indicated hsp104 mutations and control wild-type [PSI+] strain (AT3) were incubated on YPD at 30°C for four days. The results are summarized in Table 3.](/cms/asset/28a04c0a-8d5f-4c44-9a0f-5c2fb5f01f4a/kprn_a_10904060_f0002.gif)
Figure 3 Hsp104-levels in mutant strains. Wild-type [psi-] (AT1), wild-type [PSI+] (AT3), Δhsp104 and 58 missense hsp104 mutants were cultured in YPD+ade liquid medium and harvested in mid-log phase. (Upper) Anti-Hsp104 immunoblotting. (Lower) Coomassie stained internal loading control.
![Figure 3 Hsp104-levels in mutant strains. Wild-type [psi-] (AT1), wild-type [PSI+] (AT3), Δhsp104 and 58 missense hsp104 mutants were cultured in YPD+ade liquid medium and harvested in mid-log phase. (Upper) Anti-Hsp104 immunoblotting. (Lower) Coomassie stained internal loading control.](/cms/asset/7db54225-b82f-4a88-98cf-ba1a635a17c8/kprn_a_10904060_f0003.gif)
Figure 4 Tetrad analysis of hsp104 mutant phenotypes monitored by nonsense suppression of the ade1-14 strain. Heterozygous diploids from matings between indicated hsp104 mutants and wild-type [PSI+] cells (NPK151) were sporulated and their progeny were incubated on YPD at 30°C for four days. Asterisks indicate the Leu+, hsp104 mutants.
![Figure 4 Tetrad analysis of hsp104 mutant phenotypes monitored by nonsense suppression of the ade1-14 strain. Heterozygous diploids from matings between indicated hsp104 mutants and wild-type [PSI+] cells (NPK151) were sporulated and their progeny were incubated on YPD at 30°C for four days. Asterisks indicate the Leu+, hsp104 mutants.](/cms/asset/3c36c5ff-e530-48ae-a8ce-0ea6feff2304/kprn_a_10904060_f0004.gif)
Figure 5 Effect of hsp104 mutations on thermotolerance. Mid-log phase cultures of the indicated strains were incubated at 37°C for one hour to induce the heat-shock response and were then incubated for a further 20 minutes at 50°C. The viability of heat-shocked cells (right) were compared to untreated cells (left) by five-fold serial dilutions on YPD+ade agar. Cells were incubated at 30°C for two days. The results are summarized in ().
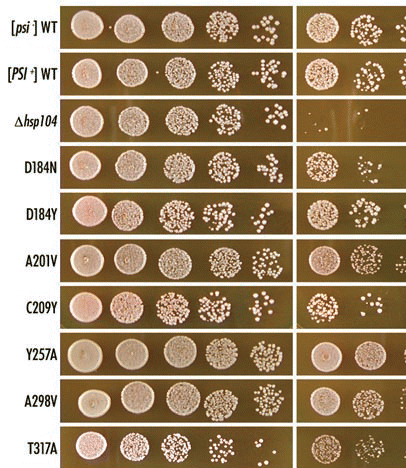
Figure 6 Structure of the ClpB/Hsp104 monomer and hexamer. (A) Ribbon drawing of T. thermophilus ClpB monomer (Protein Data Bank accession number 1QVR).Citation6 The figure shown was generated with the program PyMOL. Domain assignments were given by Lee et al.Citation6 Amino acid changes in hsp104 mutants (shown as grey, red or blue spheres) are assigned at the equivalent positions in ClpB (shown in parentheses). Red spheres show [PSI+]-eliminating thermotolerant mutations. Y257A and A298V mutations are not shown because these positions were not determined in the 3D structure of T. thermophilus ClpB. Blue spheres show hsp104 mutations that were able to propagate [PSI+] but lost thermotolerance. (B) Axial (left-top) and lateral (left-bottom) images of ClpB/Hsp104 hexamer (reprinted from Lee et al6 with permission from Elsevier). (Right) A close-up view of the lateral channel. The figure shown was generated with the program PyMOL. D184Y, A201V, T317A, L462R, P557L, D704N and T726I mutations, which remained thermotolerant but lost [PSI+] propagation, are indicated as red spheres. Parentheses indicate the equivalent positions in ClpB.
![Figure 6 Structure of the ClpB/Hsp104 monomer and hexamer. (A) Ribbon drawing of T. thermophilus ClpB monomer (Protein Data Bank accession number 1QVR).Citation6 The figure shown was generated with the program PyMOL. Domain assignments were given by Lee et al.Citation6 Amino acid changes in hsp104 mutants (shown as grey, red or blue spheres) are assigned at the equivalent positions in ClpB (shown in parentheses). Red spheres show [PSI+]-eliminating thermotolerant mutations. Y257A and A298V mutations are not shown because these positions were not determined in the 3D structure of T. thermophilus ClpB. Blue spheres show hsp104 mutations that were able to propagate [PSI+] but lost thermotolerance. (B) Axial (left-top) and lateral (left-bottom) images of ClpB/Hsp104 hexamer (reprinted from Lee et al6 with permission from Elsevier). (Right) A close-up view of the lateral channel. The figure shown was generated with the program PyMOL. D184Y, A201V, T317A, L462R, P557L, D704N and T726I mutations, which remained thermotolerant but lost [PSI+] propagation, are indicated as red spheres. Parentheses indicate the equivalent positions in ClpB.](/cms/asset/3a9103ed-ec37-411a-838a-a951dc41ac08/kprn_a_10904060_f0006.gif)
Table 1 Strains
Table 2 Primers used in this study
Table 3 Properties of hsp104 mutations