Figures & data
Figure 1 Isolation of 103Q aggregates from yeast cells. (A) Fluorescent micrograph (FITC channel) of yeast wild type cells with GFP-labeled 103Q aggregates after 6 h induction. (B) Scheme of the purification procedure. (C) Fluorescent micrographs of the indicated fractions at x100 magnification; a small portion of the immunoprecipitate (IP) was resuspended in the lysis buffer to obtain the image. (D) SDS-PAGE (Coomassie stained) gel shows approximately equal IgG distribution between IP and supernatant and almost complete purification of the IP from BSA which remains the major band in the non-precipitated fraction, but is almost excluded from the precipitate. Precipitates were reconstituted in the SDS-containing buffer in a volume equal to the volume of the supernatant.
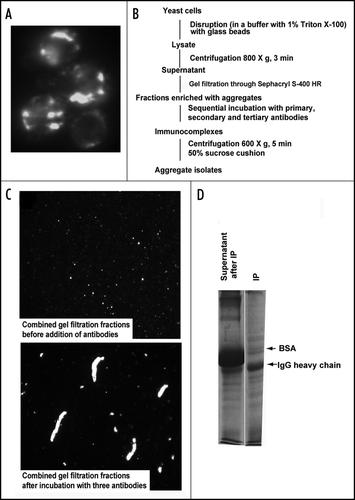
Figure 2 Isolated aggregates and control fraction analyzed by 2-D electrophoresis gels stained with Coomassie blue.
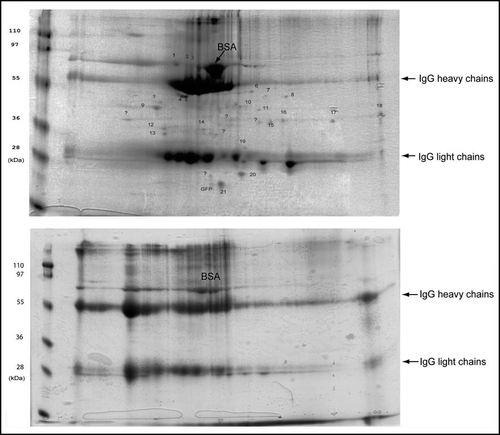
Figure 3 Mass spectra from one of the identified proteins. (A) A representative MALDI-TOF mass spectrum shows matched peptides from the identified protein, Sgt2. The sequence of the Sgt2 protein is shown on the right top corner, identified peptides are underlined. Matched peptides and the monoisotopic m/z values of fragments are indicated above each peak. (B) A representative ESI MS/MS spectrum confirms the identification of proteins by tandem MS/MS of [M+2H]2+ m/z 697.9 matching peptide m/z 1394.Citation7 of the panel a. Matched peptide sequence of Sgt2 protein is shown on the top and resulting b and y ions are indicated.
![Figure 3 Mass spectra from one of the identified proteins. (A) A representative MALDI-TOF mass spectrum shows matched peptides from the identified protein, Sgt2. The sequence of the Sgt2 protein is shown on the right top corner, identified peptides are underlined. Matched peptides and the monoisotopic m/z values of fragments are indicated above each peak. (B) A representative ESI MS/MS spectrum confirms the identification of proteins by tandem MS/MS of [M+2H]2+ m/z 697.9 matching peptide m/z 1394.Citation7 of the panel a. Matched peptide sequence of Sgt2 protein is shown on the top and resulting b and y ions are indicated.](/cms/asset/65b346df-62cf-4aa6-bf80-976fd520ee56/kprn_a_10904440_f0003.gif)
Figure 4 Colocalization of the proteins coprecipitated with 103Q aggregates. Fixed yeast cells with GFP-tagged endogenous proteins (marked on the right) and transformed with mRFP-tagged 103Q were grown in selective medium with raffinose and induced for 6 h with galactose.
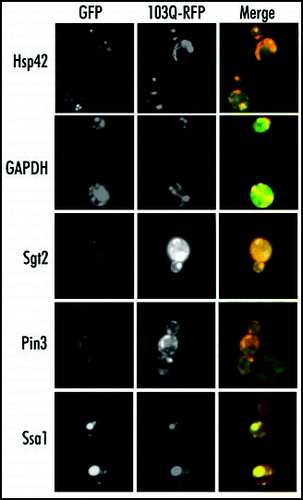
Figure 5 Isolation and analysis of 103Q aggregates from cells without prions. (A) Morphology of 103Q aggregates formed in hsp104 cells differs significantly from those in wild type cells (). (B) Fractions after gel filtration of lysates of hsp104 cells expressing 103Q analyzed by immunoblot with anti-GFP antibody, and quantified by Quantity One software (Bio-Rad). (C) Comparison of chaperones in immunoprecipitates from the wild type and mutant cells. Immunoblot with the respective antibodies (indicated on the right). To detect 103Q, true blot rabbit anti-mouse IgG was used as a secondary antibody. The aggregates were isolated from equal volumes of yeast cultures and solubilized in equal volumes of the Laemmli loading buffer.
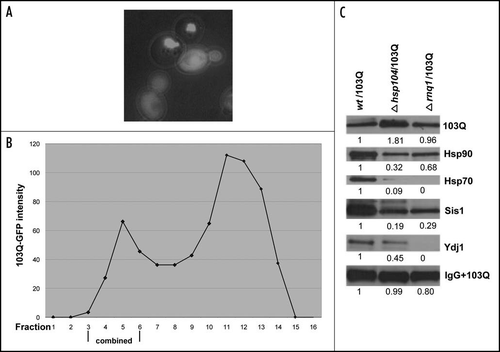
Table 1 List of identified proteins associated with 103Q aggregates