Figures & data
Figure 1 The yeast [PSI+] prion is transmitted as an extrachromosomal genetic element. The cytoplasm of [PSI+] cells contains a number of distinct physical entities—propagons—that are necessary for the continued propagation of the [PSI+] state. When a [PSI+] cell is mated to a propagon-free [psi-y] cell (left panel) the diploid is [PSI+] and all four haploid meiotic progeny are also [PSI+]. When a [PSI+] cell is mated to a [psi-] kar1cell that is defective in nuclear fusion (right panel) the resulting unstable heterokaryons generate new haploid cells—cytoductants. At a frequency approaching 100%, such cytoductants are [PSI+] irrespective of the haploid nucleus carried. NB: The nuclei arising from fusion between two parental nuclei are shown with hatches.
![Figure 1 The yeast [PSI+] prion is transmitted as an extrachromosomal genetic element. The cytoplasm of [PSI+] cells contains a number of distinct physical entities—propagons—that are necessary for the continued propagation of the [PSI+] state. When a [PSI+] cell is mated to a propagon-free [psi-y] cell (left panel) the diploid is [PSI+] and all four haploid meiotic progeny are also [PSI+]. When a [PSI+] cell is mated to a [psi-] kar1cell that is defective in nuclear fusion (right panel) the resulting unstable heterokaryons generate new haploid cells—cytoductants. At a frequency approaching 100%, such cytoductants are [PSI+] irrespective of the haploid nucleus carried. NB: The nuclei arising from fusion between two parental nuclei are shown with hatches.](/cms/asset/a6dc9539-fb8a-448e-81b5-7b926ad17dff/kprn_a_10904665_f0001.gif)
Figure 2 The molecular basis of the [PSI+]-associated nonsense suppression phenotype. Left) When a stop codon arrives at the ribosomal A site it is efficiently recognised by the release factor which in yeast and other eukaryotes is comprised of two proteins, eRF1 (Sup45p) and eRF3 (Sup35p). There are some near-cognate tRNAs able to translate termination codons albeit inefficiently and these are only usually detected when the termination machinery is impaired as, for example, is the case in [PSI+] cells or in sal mutants. Translation then proceeds to the next in-frame stop codon, which here is UAA. Right) If the cell encodes a mutant tRNA which is able to recognise a defined stop codon i.e., a cognate nonsense suppressor tRNA such as the SUQ5-encoded tRNASer 8, this tRNA is able to compete more efficiently than a near-cognate tRNA with the release factor for the termination codon at the A site. Again the efficiency with which the cognate suppressor tRNA competes greatly increased if termination machinery is impaired.
![Figure 2 The molecular basis of the [PSI+]-associated nonsense suppression phenotype. Left) When a stop codon arrives at the ribosomal A site it is efficiently recognised by the release factor which in yeast and other eukaryotes is comprised of two proteins, eRF1 (Sup45p) and eRF3 (Sup35p). There are some near-cognate tRNAs able to translate termination codons albeit inefficiently and these are only usually detected when the termination machinery is impaired as, for example, is the case in [PSI+] cells or in sal mutants. Translation then proceeds to the next in-frame stop codon, which here is UAA. Right) If the cell encodes a mutant tRNA which is able to recognise a defined stop codon i.e., a cognate nonsense suppressor tRNA such as the SUQ5-encoded tRNASer 8, this tRNA is able to compete more efficiently than a near-cognate tRNA with the release factor for the termination codon at the A site. Again the efficiency with which the cognate suppressor tRNA competes greatly increased if termination machinery is impaired.](/cms/asset/bfb54d66-e0aa-458f-9e02-c4a6a6f7374b/kprn_a_10904665_f0002.gif)
Figure 3 A genetic cross can differentiate between mutations that eliminate the [PSI+] prion (PNM, ‘PSI-No-More’ mutants) and dominant antisuppressor mutants (ASU). The diploid formed between a PNM mutant and a [PSI+] strain loses the ability to replicate the [PSI+] prion because it can no longer efficiently produce the new propagons required. Consequently all of the haploid meiotic spores do not inherit the ability to propagate the [PSI+] state i.e., are [psi-]. In contrast, in a cross between an ASU mutant and a [PSI+] strain, although the phenotype is the same as the PNM/+ diploid, the ASU/+ diploid still has the ability to generate new propagons. The reason the nonsense suppression phenotype is not expressed is because these cells also produce a significant level of soluble and functional Sup35p which ensures that efficient translation termination occurs. Consequently those meiotic spores that inherit the ASU mutation show a red nonsuppressed phenotype whereas those that do not, show the [PSI+] phenotype.
![Figure 3 A genetic cross can differentiate between mutations that eliminate the [PSI+] prion (PNM, ‘PSI-No-More’ mutants) and dominant antisuppressor mutants (ASU). The diploid formed between a PNM mutant and a [PSI+] strain loses the ability to replicate the [PSI+] prion because it can no longer efficiently produce the new propagons required. Consequently all of the haploid meiotic spores do not inherit the ability to propagate the [PSI+] state i.e., are [psi-]. In contrast, in a cross between an ASU mutant and a [PSI+] strain, although the phenotype is the same as the PNM/+ diploid, the ASU/+ diploid still has the ability to generate new propagons. The reason the nonsense suppression phenotype is not expressed is because these cells also produce a significant level of soluble and functional Sup35p which ensures that efficient translation termination occurs. Consequently those meiotic spores that inherit the ASU mutation show a red nonsuppressed phenotype whereas those that do not, show the [PSI+] phenotype.](/cms/asset/d1266962-72d3-4dd7-9e98-36a8d84f877e/kprn_a_10904665_f0003.gif)
Figure 4 Mutations within the molecular chaperone Hsp104 can give rise to a ‘PSI-No-More’ phenotype. Upper: A schematic representation of the key functional domains of Hsp104 showing the two nucleotide binding domains (ATP; NBD1/NBD2) and the N-terminal (NTD) and C-terminal (CTD) domains plus the linker region between the two NBDs. The proposed functional roles of the different domains are indicated below. See refs. Citation45 and Citation46 for further discussion on the functional organisation of Hsp104. Lower: mutations that have so far been described which give a ‘PNM’ phenotype. The location of the mutations is given together with an indication of whether the mutation in question is dominant or a recessive with respect to the ‘PNM’ phenotype. ND indicates that the dominance/recessive character of the mutant was not reported.Citation1 The single mutations K218T and K620T are also dominant/semi-dominant PNMs in some genetic backgrounds.
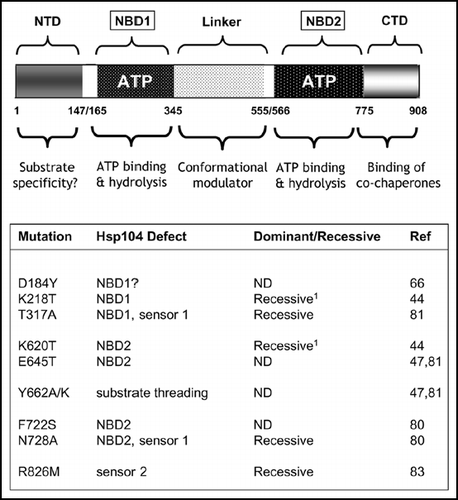
Figure 5 The prion-forming domain of the Sup35p protein. (A) The 685 residue Sup35p protein has three regions defined by the locations of the first three in-frame AUG (Met) codons: the amino terminal N domain (residues 1–123) that is absolutely required for the prion behaviour of the protein; the highly charged middle region (M; residues 124–253) and the carboxyl-terminal domain (C; residues 254–685) which carries the essential release factor activity.Citation20,Citation48 (B) Within the N domain lies a region, the so-called prion-forming domain-which contains a region important for aggregation and a separate but overlapping region important for propagation of the prion state.Citation48 The aggregation element contains a short peptide sequence based around residues 7 to 13, that is highly amyloidogenic. The propagation element contains five imperfect copies of an oligopeptide repeat sequence (R1–R5). (C) The ‘PSI-No-More’ mutant PNM2-1 carries a single amino acid substitution (Gly to Asp) within repeat R2 of the propagation element. This single substitution leads to a form of Sup35p that can still aggregate but can no longer be efficiently propagated in most laboratory strains.Citation20,Citation48,Citation50
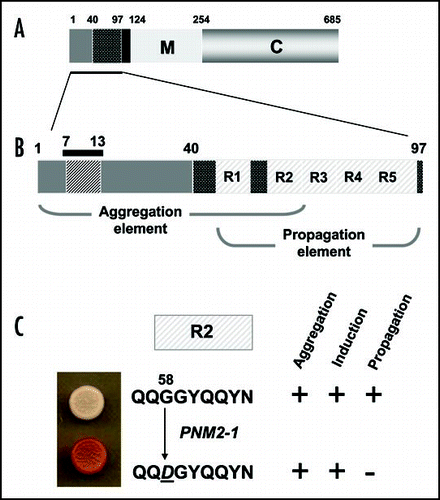
Table 1 Nuclear genetic antagonists of the [PSI+]-associated nonsense suppression phenotype