Figures & data
Figure 1 Overview of the PrP sequence. (A) Block diagram of the PrP architecture. The residue numbering refers to huPrP. The positions of the glycosilation sites are indicated at Asn181 and Asn197. Cys179 is covalently bound to Cys214. The number of octarepeats depends on the species considered. (B) Charge distribution along the sequence of huPrP. Only the charged residues are indicated. The N-terminus contains only positively charged residues (11 charges), whereas the C-terminus contains both types, with a slight excess of negative residues.
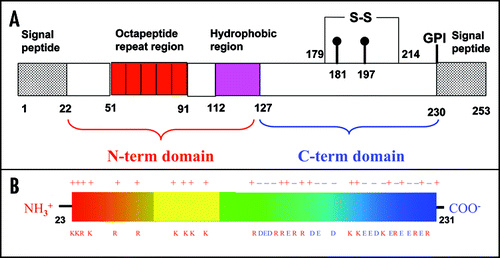
Figure 2 Sequence alignment and overall fold of PrP. (A) Sequence alignment of a subset of PrP precursor sequences chosen as the ones for which the structure of the C-terminal domain is available (boxed in blue). The alignment was obtained and color coded by Clustalx.Citation67 Stars, semicolons and dots refer to conserved and partially conserved residues, according to Clustalx convention. (B) Ribbon representation of the C-terminal domain of mPrP.Citation68 The secondary structure elements and the N- and C-termini are labelled. The sulphur bridge is indicated in yellow.
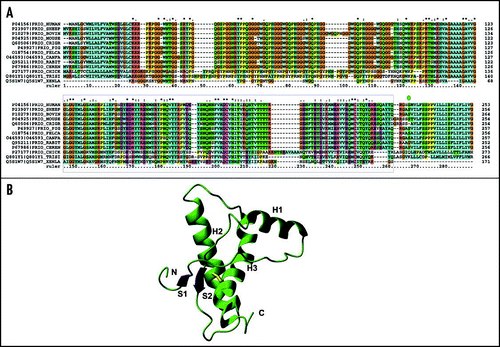
Figure 3 Bundles of the NMR structures. (A) Superposition of the nine huPrP structures. The backbone atoms of the secondary structure elements (residues 129–130, 145–152, 162–163, 173–190, 200–222) were arbitrarily superposed on the coordinates of 1qm0 (shown in red). The minimised structure was used when available. Otherwise, the first structure of the bundle was selected. (B) Superposition of the PrP structures from other mammals. The best pairwise superposition according to Dali was found arbitrarily using the coordinates of mPrP (shown in red). (C) The coordinates of the C-terminal domains of bird and reptile PrPs. Left, turtle; middle, frog; right, chicken. The same orientation as in Figure 2B was chosen.
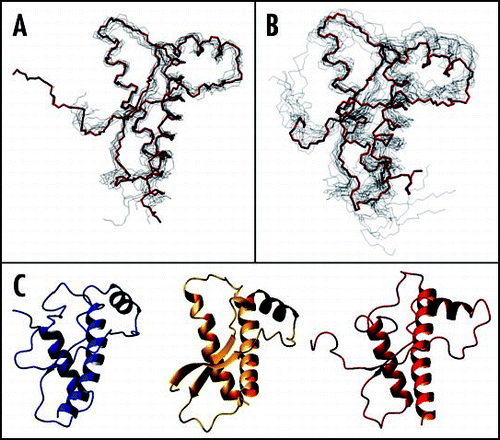
Figure 4 Comparison between the X-ray structures of the PrP C-terminal domain. (A) Superposition of shPrP (1uw3, in red) from Leu125 to Ala230 (huPrP numbering) with shPrP (1tpx, in blue) from Gly127 to Tyr228. (B) Superposition of shPrP (1uw3, in red) from Leu125 to Ala230 (huPrP numbering) with huPrP (1i4m, chain A in cyan, chain B in magenta) from residue Gly119A to Thr191A and from residue Glu196B to Tyr226B.
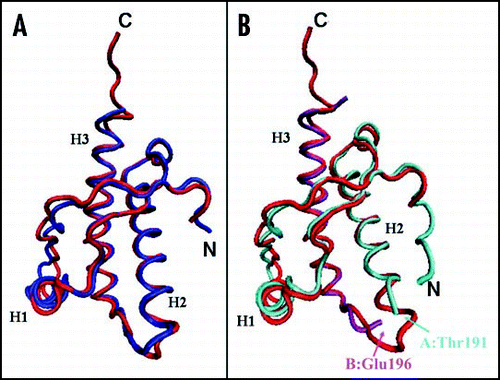
Figure 5 Schematic representation of the fold of the PrP C-terminal domain, indicating the most relevant ionic interactions (within 3.8 Å) as observed in the X-ray structures. The disulfide bridge connecting H2 to H3 is indicated in yellow. Residue numbering, sequence (apart from the mutation R148C) and secondary structure assignment refer to huPrP and are consistent with 1uw3 (the sequence of the antibody-bound shPrP differs by +3). Ellipses represent residues in α-helix; green rectangles represent residues in β-strand; diamonds represent residues in 310 helix; transparent rectangles represent charged residues in loops. Red, blue, green, purple and cyan lines refer to interactions observed in the 1u3w, 1tpx, 1tqb, 1tqc and 1i4m structures, respectively. Interactions within the dimer were considered for 1i4m. Inter-molecular interactions between adjacent molecules in the crystal state are shown by red dotted lines.
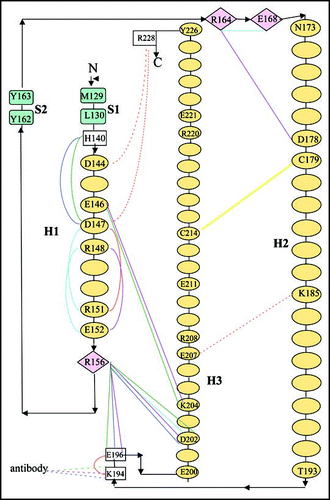
Figure 6 Interfaces observed in the crystal structures (see also ). (A) Dimeric swapped structure of huPrP (1i4m). Chains A and B are drawn in cyan and magenta respectively. The extended surface partly encompasses H1, H2, H2' and H3. A detailed view of the hinge region between the two subunits is shown in the inset. The newly formed β-strand encompasses the polyThr stretch (190–194). (B) shPrP structure (1uw3). The two symmetry axis related molecules are drawn in red and green. The contact surface encompasses the C-terminus of H2, the connecting loop, and the N-terminus of H3. The ionic interaction between the side chain of Lys185 and Glu207 is indicated. H-bonds close to the surface are omitted for clarity. (C) Close up view of shPrP 1uw3 showing how the β-sheet elongates through interaction between adjacent molecules. The side chains of selected residues are indicated. Met129 and Leu130 are labelled only once. Only backbone H-bonds are drawn, with N and O atoms indicated by spheres. (D) β-sheet elongation in the 1tpx shPrP structure (in blue). The antibody C chain is drawn in green. The contact surface encompasses PrP S1 and the short edge strand belonging to the four-stranded β-sheet, named I, of the antibody C chain. Only two strands of sheet I are drawn.
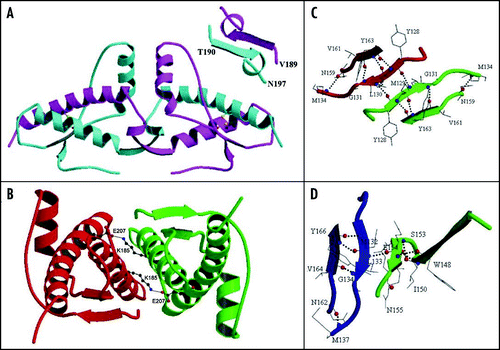
Figure 7 Conserved hydration site in the crystal state. The figure refers to shPrP (1uw3), as an example. Distances are given in Å.
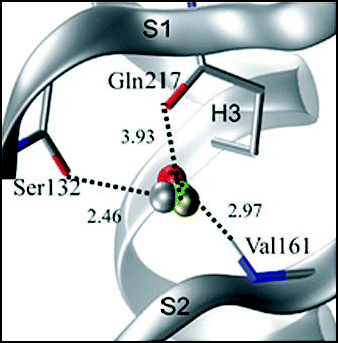
Figure 8 Structural comparison between the C-terminal domain of PrP and Dpl. The structures of (A) mPrP (1ag2), (B) hamster PrP (1b10), (C) mouse Dpl (1i17) and (D) human Dpl (1lg4) were superposed by the Dali server and then displaced. The two Dpl structures superpose with an r.m.s.d. of 2.0 Å over 97 residues (Z-score 13.45), whereas the two mouse paralogs superpose with an r.m.s.d. of 3.8 Å over 87 residues (Z score 5.0).
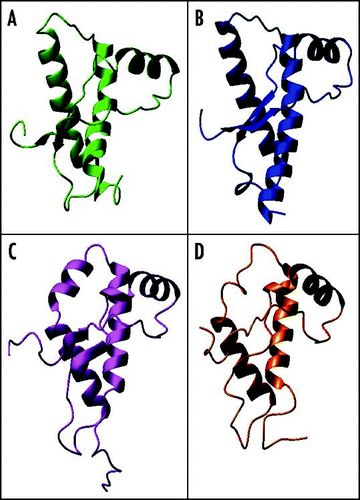
Figure 9 Current models of PrPSc. (A) Trimeric model of PrP27–30 after Govaerts et al., 2004.Citation55 (B) Crystal structure of the PrP SNQNNF peptide (PDB code 2ol9).Citation124
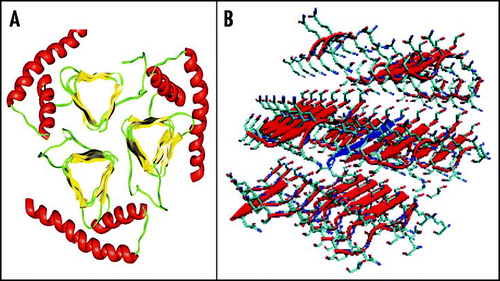
Table 1 Summary of relevant information about the NMR structures
Table 2 Summary of relevant information about the X-ray structures
Table 3 R.m.s.d. (Å) between the X-ray and the NMR structures; the number of the Cα atoms superposed is given in parentheses
Table 4 Classification of a few selected interfaces observed in the crystal state; only some interfaces with an area ≥380 Å2 were listed