Figures & data
Figure 1 Structural organization of prion proteins. QN: the QN-rich stretch. OR: the region of oligopeptide repeats. PrD-prion domain. Numbers correspond to amino acid (aa) positions. Arrows indicate domain and subdomain boundaries. N, M and C-N-proximal, middle and C-proximal regions of Sup35, respectively. The N/M and M/C boundaries are arbitrarily assigned to the second (aa 124) and third (aa 254) methionine residues of the Sup35 protein. See text for details.
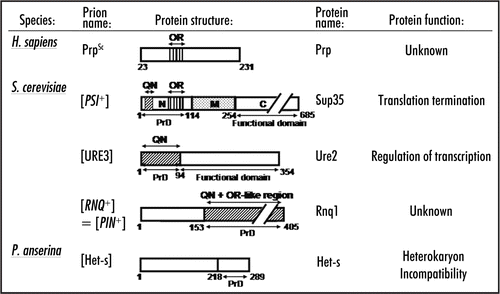
Figure 2 Evolutionary comparison of the N-terminal domains of Sup35 homologs. Sequences are from www.ncbi.nlm.nih.gov. Taxonomical relationships are from www.ncbi. nlm.nih.gov/Taxonomy. Scales do not correspond to evolutionary distances. For QN and OR designations, see . Numbers on the right correspond to the size of the N-terminal region (in aa) in each case. Sequence data were obtained from www.ncbi.nlm.nih.gov. ? -refers to the cases where search for prion activity in S. cerevisiae has been performed but have not yielded positive results (O. Zemlyanko, A. Petrova and G. Zhouravleva, unpublished; K. Gokhale and Y. Chernoff, unpublished). NT, not tested.