Figures & data
Figure 1. (A) Schematic of the α-hemolysin pore embedded in a lipid membrane. Unfolded and simple α-helical or β-sheet forming peptides can readily translocate. (B) Typical current trace of Aβ1−40 recorded for 10 sec. The open pore current is 100 pA and each spike represents an event where a single peptide interacts with the pore. Typically for a peptide, large spikes are due to translocations and short spikes are bumping events.
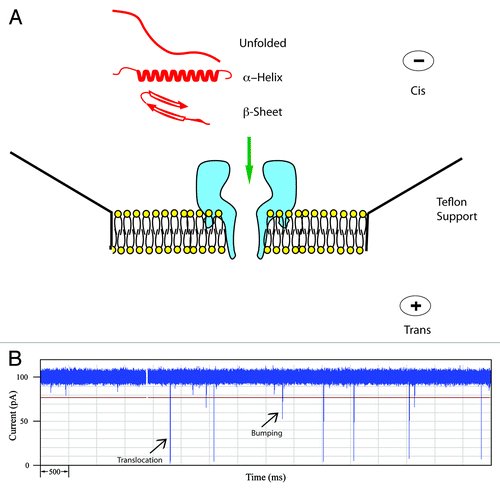
Table 1. Peptides and proteins used in this work
Figure 2. Event histograms for Aβ peptides. Blockade current and blockade time histograms for Aβ1−42 (A) and (B), Αβ1–40 (C) and (D), and mutant Aβ1–40 D23N (E) and (F). Stock solutions of peptides (rPeptide, Bogart GA) were dissolved in 50% TFE at 1 mg/ml and 10 μL was added to the cis-side of the nanopore chamber which contained 1 M KCl, 10 mM HEPES, pH 7.8. The applied voltage was 100 mV. Further details of the experimental set up have been described previously.Citation12,Citation20-Citation22 (See also Supplementary Information).
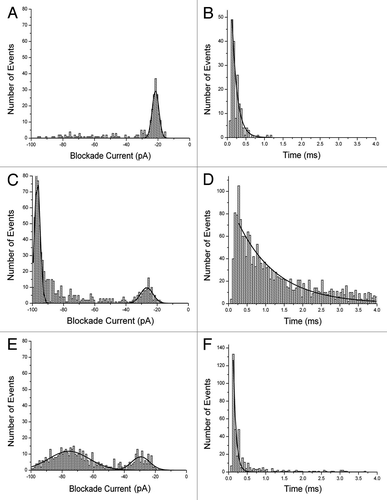
Figure 3. Blockade current histograms for α-synuclein. (A) wild type, (B) mutant A30P, (C) mutant E46K and (D) mutant A53T. The peptides (rPeptide) were dissolved in 10 mM TRIS-HCl, pH 7.4 at 1 mg/ml and 10 μL was added to the cis-side of the nanopore chamber which contained 1 M KCl, 10 mM HEPES, 1 mM EDTA, pH 7.8. The applied voltage was 100 mV.
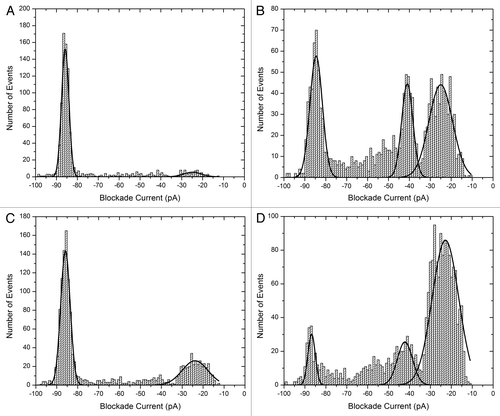
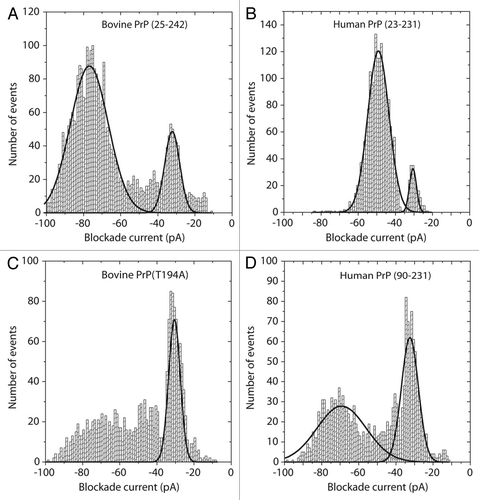
Figure 4. Blockade current histograms for prion proteins. (A) Bovine PrP (25–242), (B) human PrP (23–231), (C) mutant bovine PrP (T194A) and (D) Human PrP (90–231). The proteins were obtained from Jena Bioscience, and dissolved in 10 mM TRIS-HCl, 0.1 mM EDTA pH 8.0 at 1 mg/ml. Twenty μL was added to 8.6 μL of 5 M Guanadinium-HCl and incubated at 21°C for 1 h. The whole aliquot was then added to the cis-side of the nanopore chamber which contained 1 M KCl, 10 mM TRIS-HCl pH 7.8. The applied voltage was 100 mV