Figures & data
Figure 1 Nucleotide and amino acid database consensus sequence and polymorphisms observed in Illinois white-tailed deer. Numbers indicate the nucleotide or deduced amino acid sequence from a consensus. Observed frequency (# of polymorphic alleles/total # of alleles* 100) of polymorphisms and domains within the prion protein are also indicated.
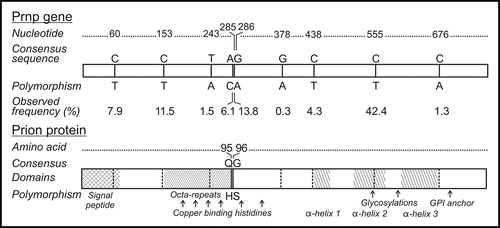
Figure 2 Probability of CWD as predicted by the number of polymorphic alleles. Total polymorphisms, silent polymorphisms, coding polymorphisms and total polymorphisms omitting locus 153 were used as independent variables in separate logistic regression models with CWD as the outcome variable. Odds ratios were estimated using the number of heterozygous and homozygous SNP summed across all loci. Coding for locus 153 was reversed because wild-type alleles conferred susceptibility (2 = homozygous for the consensus genotype, 1 = heterozygous, 0 = homozygous for the polymorphism), which resulted in a higher predicted probability of CWD than in a model omitting locus 153.
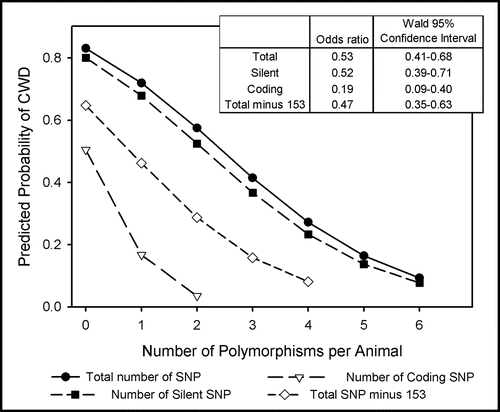
Figure 3 Distribution of coding polymorphisms Q95H and G96S. Bars represent the percent of sampled population of the indicated genotype within disease status. Distribution of coding polymorphisms for sampled populations of Illinois deer are represented by open bars. Distribution of polymorphisms for Wisconsin deer (hatched bars) were calculated from published results.Citation12
Table 1 Ratio of observed and expected allele frequencies for nucleotide polymorphisms
Table 2 Ratio of observed and expected genotype frequencies for nucleotide polymorphisms
Table 3 Reconstructed frequencies of common haplotypes for CWD positive animals and controls