Figures & data
Figure 1 A proposed model for the formation of IDE-AβSCx. Monomeric Aβ (Aβm) may follow a pathway of self-assembly to form oligomers (Aβo) or interact with the binding site (or at least a part of it) of fully active or inactive IDE conformers (IDEa and IDEi, respectively) to form a reversible IDE-Aβ encounter complex. IDEa and IDEi may be or not in equilibrium. IDE-AβSCx formation proceeds very slowly (represented by the dashed arrows) as an irreversible process that involves a conformational change in both molecules. In the case of IDEa-Aβ at neutral pH, the pathway is largely favored toward proteolysis of Aβ and free IDE (not depicted). Alternatively, it may lead to the slow formation of IDE-AβSCx.
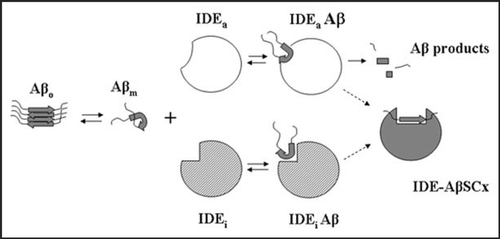
Figure 2 Endogenous IDE-AβSCx from rat brain. Rat brain IDE was immunoprecipitated with monoclonal anti-IDE 3A2, proteins resolved on SDS-PAGE and the 120 kDa specific band digested in-gel with endo-LysC. Products were analyzed by MALDI-TOF as describedCitation13 and 51 peaks were assigned to IDE (not shown). The depicted spectrum shows a peak (peak A) with the average mass of rodent Aβ1–28 (error <1 dalton), a predicted product of endo-Lys C digestion of Aβ between Lys28-Gly29 (inset). Obs. M, observed mass; Calc. M, calculated mass. This peptide did not match to IDE sequence and was not found after digestion of a piece of gel from the 120 kDa region of a rat brain inmunoprecipitation with an unrelated antibody (not shown).
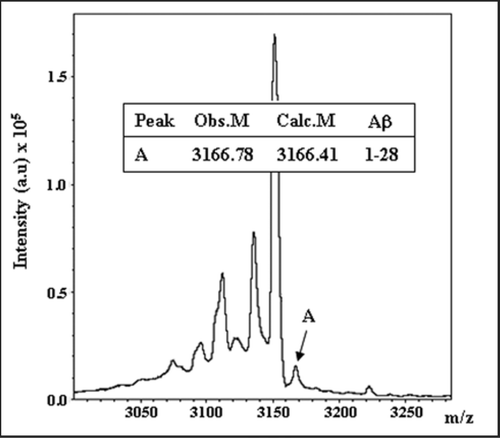
Figure 3 Proposed IDE-AβSCx formation and fate in a cellular context. IDE may interact with extracellular Aβ or with newly generated Aβ along the endocytic pathway. At neutral pH, IDE retains full catalytic activity and degrades Aβ monomers. As acidification increases along the pathway, IDE activity is reduced and more IDE-AβSCx is generated. Unbound monomeric Aβ, in turn, may aggregate readily favored by its isoelectric point. The formation of IDE-AβSCx may trap Aβ monomers irreversibly, preventing their massive oligomerization in acidic compartments. Possible destinations of IDE-AβSCx include its translocation and targeting to the proteasome, lysosomal degradation or secretion via exosomes. MVB, multivesicular bodies. The triangles on the right side depict schematically the opposite trends of IDE activity and Aβ aggregation within the intra-luminal pH range indicated by the numbers beside subcellular organelles (reviewed in ref. Citation40). For clarity, the more abundant IDE in the cytosol and other possible sites of Aβ aggregation are not depicted.
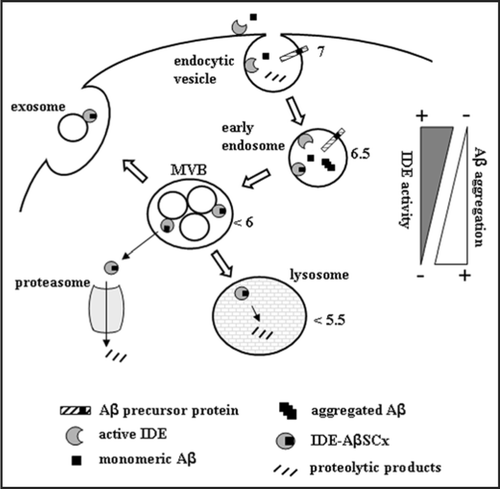
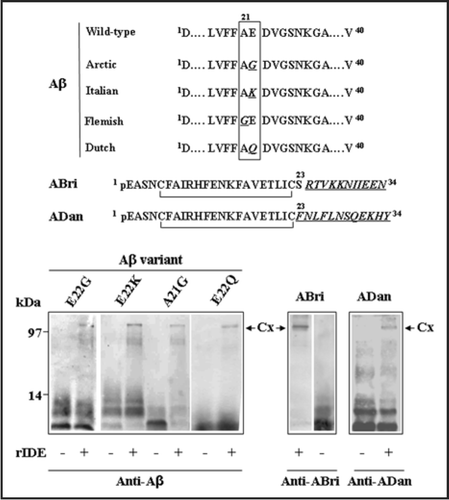
Figure 4 Stable complexes formed by IDE with different amyloid peptides. Recombinant rat IDE (rIDE) was incubated at a 25:1 molar ratio (peptide:enzyme) for 3 h at 37°C with the indicated amyloid peptides. Aβ1–40 genetic variants containing amino acid substitutions are associated with hereditary forms of AD or amyloid angiopathy and stroke. The ABri and ADan peptides are associated with familial British and Danish dementia, respectively. Top, schematic representation of the variant peptides with the amino acid substitutions in italics and underlined (the one-letter code is used). pE denotes pyroglutamic acid. The C-termini of ABri and ADan are the result of a loss of a stop codon after Ser23 or a duplication-insertion immediately before the stop codon, respectively. The N-termini are stabilized by intra-chain disulfide bonds. Bottom, Western blots with monoclonal anti-Aβ 6E10 or polyclonal anti-ABri or anti-ADan to detect Aβ, ABri or ADan peptides alone or in complex with rIDE, respectively. Before running the gel, samples were boiled in 8% SDS containing 0.1 M DTT for 5 min. The stable complexes are indicated by arrows (Cx). On the left, molecular mass standards, in kilodaltons.