Figures & data
Figure 1 R1 and R5 play important roles in guiding CsgB-responsiveness and CsgA seeding. (A) A schematic of the CsgA primary structure, including a Sec signal sequence (which is cleaved after translocation across the cytoplasmic membrane), N-terminal 22 residues involved in secretion through the outer membrane and five repeating units comprising a protease-resistant amyloid core. The regularly spaced Ser, Gln and Asn residues in each repeating unit are enclosed in boxes. (B) A summary of the interactions between CsgA and CsgB (heteronucleation), or CsgA derivatives (seeding). A “+” means that the interaction reduces the CsgA lag phase. A “−” means that the interaction has little or no affect on the CsgA lag phase. CsgA seeding was measured as previously describedCitation11 and CsgB heteronucleation was measured by the overlay assay.Citation10 (C) Phylogram of consensus sequences (SerX5GlnX4AsnX5Gln) of five repeating units. The sequence similarity and identity between consensus sequence of R3 and R5 are higher than those between R1 and R5. (D) The nucleation model of CsgA in vivo polymerization. Curli assembly is governed by surface-localized CsgB (heteronucleation) and by the growing fiber tip (homonucleation). CsgG is the major component of the curli secretion apparatus that directs CsgA and CsgB across the outer membrane. After secretion from the periplasm to extracellular space, R1 and R5 of CsgA can interact with CsgB or fiber tips, initiating curli formation.
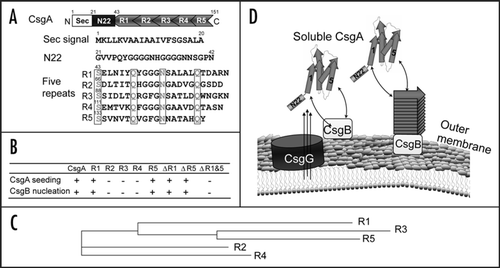
Figure 2 Salmonella typhimurium CsgA responds to Escherichia coli CsgB in E. coli ΔcsgA cells and assembles into curli. (A) The YESCA plates supplemented with Congo red dye with E. coli ΔcsgA cells transformed with vector control or plasmids encoding E. coli CsgA or S. typhimurium CsgA. Cells were grown for 48 hrs at 26°C. (B) The Western blot of E. coli ΔcsgA cells transformed with vector control (lanes 3 and 4) or plasmids encoding E. coli CsgA (lanes 1 and 2) or S. typhimurium CsgA (lanes 5 and 6). Samples were treated with (+) or without (−) formic acid (FA) to depolymerize the polymers. The blot was probed with anti-CsgA antibody. (C) Schematic shows cross nucleation and polymerization occurring between different bacterial species in the same environment. CsgA molecules secreted from species I (indicated in blue) interact with CsgB of Species II (indicated in red), undergo a conformation change and assemble into hybrid fibers (indicated in orange). Species I and II can assemble their own fibers (indicated in blue and red, respectively).
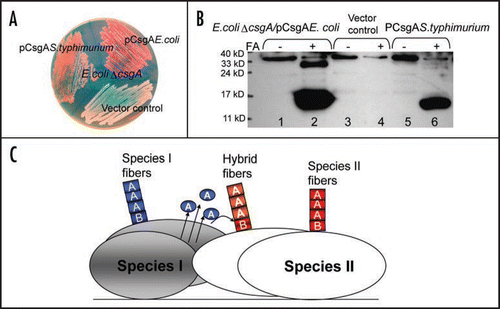
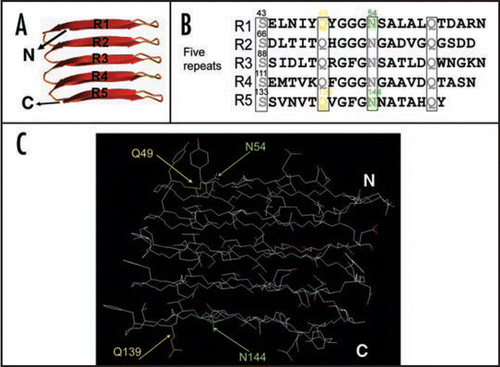
Figure 3 Critical side chains in curli assembly. (A) The predicted structure of CsgA in curli structure. The structure was constructed by Dr. Craig Smith based on previous work from the Kay lab.Citation20 (B and C) The regularly spaced Ser, Gln and Asn residues in repeating units are enclosed in four boxes and their roles in curli assembly were analyzed by Ala substitutions. The critical residues Q49, N54, Q139 and N144 were highlighted in yellow (for Gln residues) and green (for Asn residues) in the primary structure of CsgA (B) and in the predicted 3D structure of five repeating units (C).