Figures & data
Figure 1 Binding of apt #1 to bPrP and bPrP-β. Binding curves of apt #1 to bPrP and bPrP-β are shown by closed circles and open circles, respectively. Solid and dashed lines represent different buffer conditions: 10 mM K+ and 100 mM Na+, respectively. The binding data are analyzed by GraphPad PRISM (see Materials and Methods).
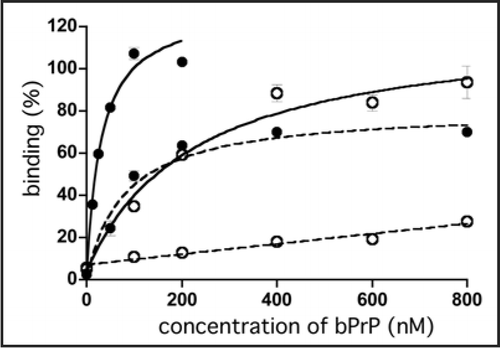
Figure 2 Detection of bPrPC in bovine brain homogenate and dot-blotting assay using a biotinylated apt #1 (Bi#1). (A) Bovine brain homogenate was analyzed using SDS-PAGE and subsequently with Northwestern blotting. bPrPC (lane 1 and 2) was detected using Bi#1 (left) and T2 antibody (right). Lane M represents protein molecular weight markers. N, M and D indicate non-, mono- and di-glycosilated PrP isoforms, respectively. (B) Dot-blotting assay of recombinant bPrP using Bi#1 or BiC#1. Number 1 represents cross-linked 5′-biotinylated RNA (Bi#1, BiC#1). Numbers 2–6 represent decreasing concentration of mounted bPrP (ng) as follows: 2 = 480, 3 = 240, 4 = 120, 5 = 60, 6 = 30.
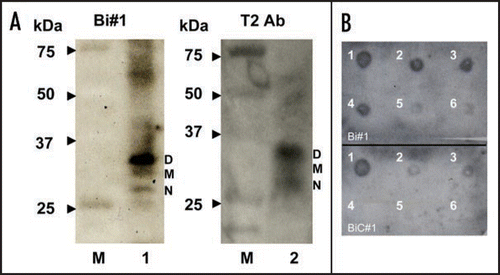
Figure 3 CD spectra of apt #1 and r(GGA)4-15 in the presence of KCl. CD spectra were measured at 20°C in titration with KCl (0, 1, 10, 100 mM). (A) CD spectra of 10 µM apt #1. (B) The structure of d(GGA)4.Citation24
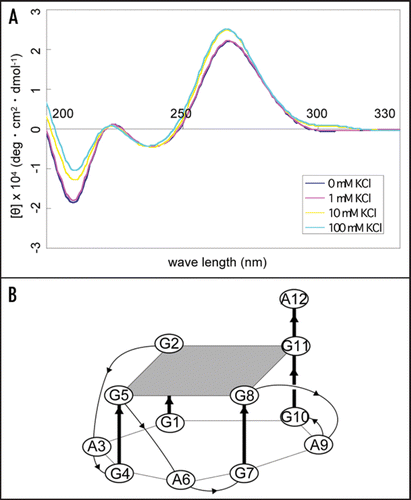
Figure 4 Binding affinity of anti-bPrP aptamer apt #1 for deletion variants of bPrP. (A) Circles, triangles and squares indicate full length bPrP (25–241), bPrP (102–241) and bPrP (132–241), respectively. (B) The amino-acid sequence alignment of bPrP. The apt #1 binding region is underlined.
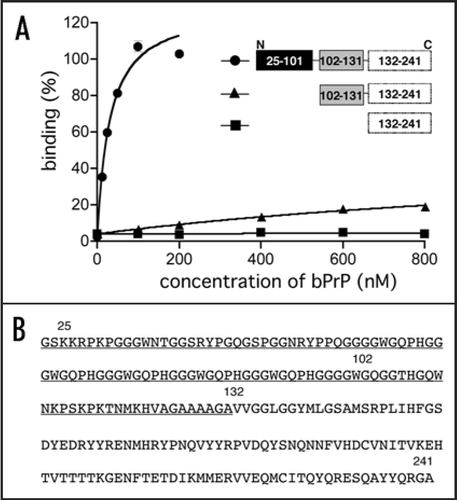
Figure 5 Comparison of binding of apt #1 and bPrP using competitive binding assay. The #1 mutants (m7, m8) contained substitutions (shown underlined) in the conserved region (GGA)4 as follows: m7: 5′-GGUGGAGGAGGA-3′; m8: 5′-GGUGGUGGUGGU-3′. The competitor RNAs were added in various concentrations with 0, 1, 5 and 10-fold greater molar concentrations of labeled apt #1. Binding assay was performed in the presence of 10 mM KCl (A) and in the absence of KCl (B).
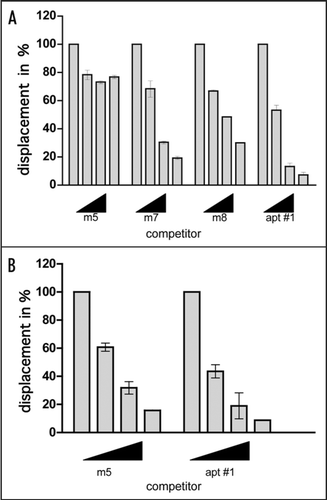
Table 1 In vitro selection conditions for recombinant bPrP
Table 2 Sequences of randomized regions and majorities of isolated RNA aptamers against recombinant bPrP
Table 3 Sequences and binding affinities of different mutants
Table 4 Comparison of binding affinities for bPrP and bPrP-β between GGA repeat containing RNAs and DNA
Table 5 Comparison of adenine stretch at 5′-site of (GGA)4 with binding affinities of bPrP, bPrP(102–241) and bPrP-β