Figures & data
Figure 1 30 Å resolution electron density map of in vitro grown mammalian prion protein fibril from ref. Citation9 (grey) with model repeat unit of four PrP proteins adopting beta helical conformations in both the N-terminal and C-terminal regions embedded (lavender). Embedding of the tetramer model and image production through Chimera.Citation65
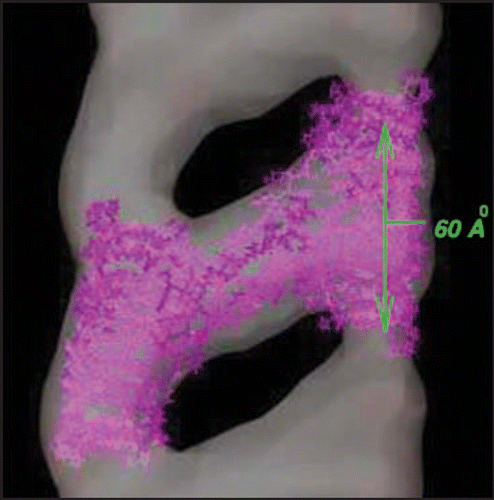
Figure 2 Comparison of PrP with LHBH structure. On the left we show human PrP (adopted from PDB structure 1QM0,Citation31) with three color coded regions: residues 90–145 (orange), residues 146–167 (green), and residues 168–230. In the LHBH structure, which is the building block for tetramers (like that shown in ), residues 90–145 go into a LHBH (N3, after ref. Citation7), 146–167 into a loop, and 168–230 into another LHBH (C4, present work).
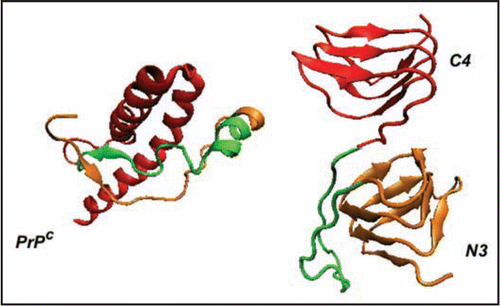
Figure 3 Index scheme for one turn of a Type II LHBH.Citation11 The nomenclature here follows reference Citation7. Note that L3,L5 point inwards.
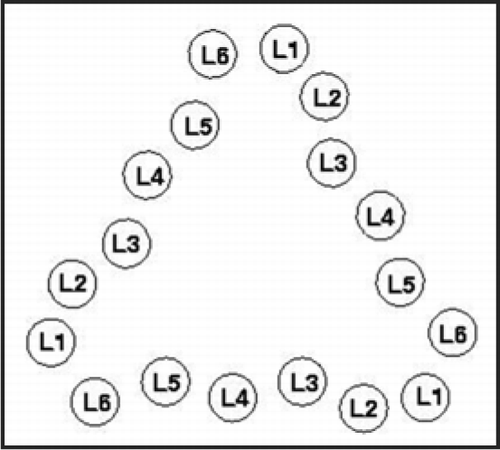
Figure 4 Threads for model β helices discussed in this paper. A single turn with 18 positions is used in the columns. Cysteines are colored gold, asparagines which link to sugars are colored blue, inward pointing acidic or basic residues are painted red. Asterisks denote loops as detailed at the bottom of the Figure, and capital X's denote jumps.
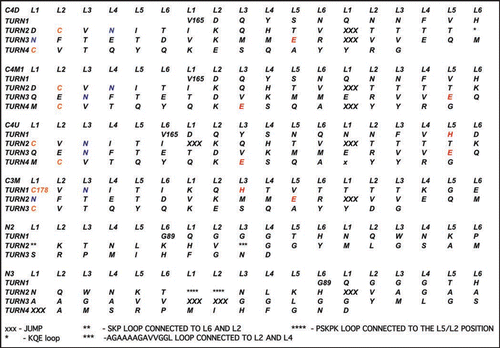
Figure 5 Left handed β helical structures for model prion tetramers. C4: 4 turn β helix for the C-terminal region (residues 166–226). Note, the cysteines highlighted with yellow spheres, glutamates highlighted in red, and glycans linked asparagines highlighted in green. N3: 3-turn β helix for the N-terminal region drawn from references Citation7 and Citation11. N2: New 2-turn N-terminal beta helix. Images produced with VMD.Citation67 Note: PSKPK denotes the small hinge region hypothesized to link the domain swap between tetramers; G-A represents the hydrophobic loop removed from N3 to stabilize N2.
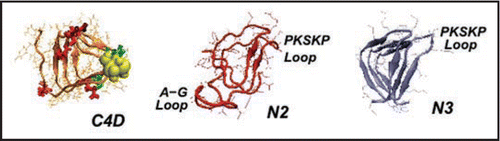
Figure 6 Different C-terminal β-helical threads accommodating 0 or 1. Depending upon the positions of the N-linking asparagines (pointing in or out, highlighted in green) the C-terminal β helix can accommodate 0 (C4U),1(C4M1) or 2 sugars(C4D, c.f. ). Cysteines are highlighted in yellow, glutamates in red.
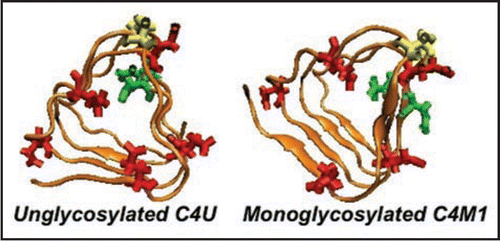
Figure 7 Root-mean square deviations (RMSD) vs. all atom AMBER8 molecular dynamicsCitation64 simulation time to 10 ns from starting structures for LHBH regions of two known proteins (1MR7—Streptogramin A AcetyltransferaseCitation68 and 1KGQ—Tetrahydrodipicolinate N-SuccinyltransferaseCitation69—and two model LHBHs (C4D—diglycoslyated and C4D-D(0)—diglycosylated and protonated glutamate), as shown in . In runs up to 1 ns which include five other known LHBH structures we obtain similar RMSDs all bracketed by 1MR7 and 1KGQ. Up to 1 ns, we find similar results for C4M1,C4M2, and N2 (not shown-order 2.5 Å), while the N3 RMSD is much larger (≈6 Å).
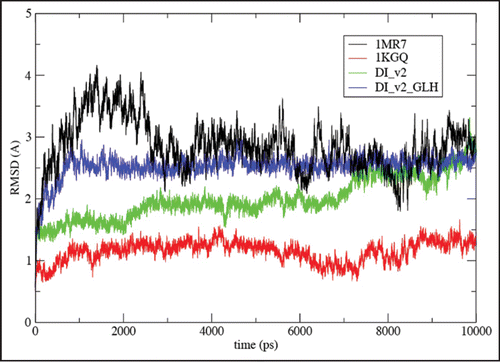
Figure 8 Empirical stability measures for known LHBHs and four model LHBHs. We compare side-chain-to-side-chain hydrogen bonding, volume packing fraction and frustration index (see text for definitions), with a positive frustration index indicating good exposure of hydrophilic residues and burial of hydrophobic residues. The C3,C4 models compare favorably to known left handed LHBH values; N3 does badly on packing and side-chain-to-side-chain hydrogen bonding, while N2 fares better. In addition to 1KGQ and 1MR7 (c.f. ) we have compared to 1SSM (Serine AcetyltransferaseCitation71), 1T3D (Serine AcetyltransferaseCitation72), 1G97 (N-acetylglucosamine-1-phosphate uridyltransferaseCitation73) and 1LXA (UDP-N-acetylglucosamine acyltransferase74).
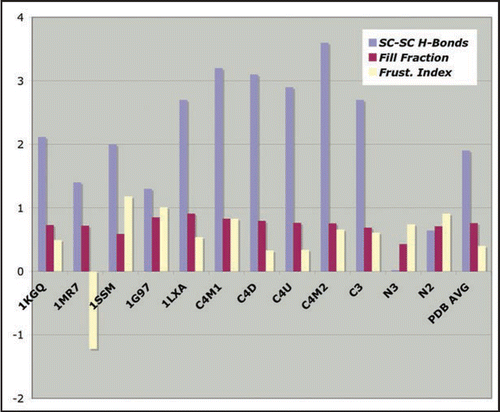
Figure 9 Schematics of proposed tetramer repeat units. Boxes: C- or N-terminal LHBHs viewed from the side (perpendicular to the helix axis). The arrow denotes the n-terminal to c-terminal progression of the sequence. Lines: Large loop regions (residues 145–166 for N2-C4 pairing, or 145–176 for N3-C3 pairing). Lines and boxes are color coded by monomer. The first row contains filaments with the same N-C sense, while the second row contains filaments with opposite N-C sense.
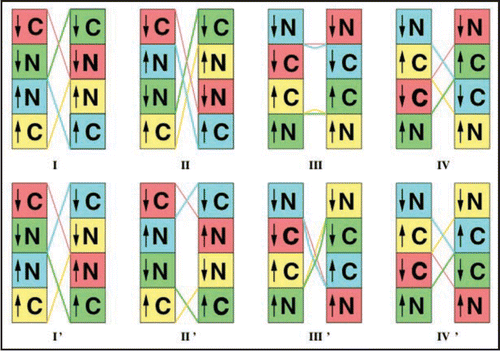
Figure 10 Schematic alignment of M129 with C-terminal LHBH for WT human, FFI human and canine prions. For WT, one S-amide hydrogen bond (with H177) is possible. For FFI, two S-amide hydrogen bonds (with H177 and N178) are possible. For canines (referenced to the human sequence) within the Swiss PDB viewerCitation66 rotamer library, no orientation of the R177 could produce amide hydrogen bonding with M129 sulfur. In each case, alignments are produced within the Swiss PDB viewerCitation66 by sweeping through low score rotamers and images via VMD.Citation67 We identify the small PSKPK loop for domain swapping between tetramers, and the hydrophobic G-A loop pulled from the N2 Model to stabilize the LHBH.
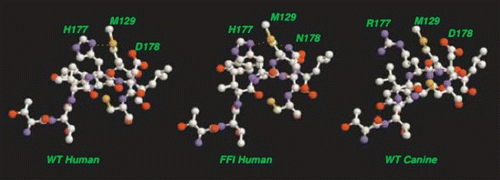
Table 1 Comparison of eight fibril repeat unit models shown in