Figures & data
Figure 1 Amino acid sequences of prion domains of known prions. Asparagine (N) residues are highlighted in blue, glutamine (Q) residues are highlighted in purple, and serine (S), tyrosine (Y) and threonine (T) are highlighted in red. Underlined areas indicate the imperfect oligopeptide repeats in Sup35 and Rnq1.
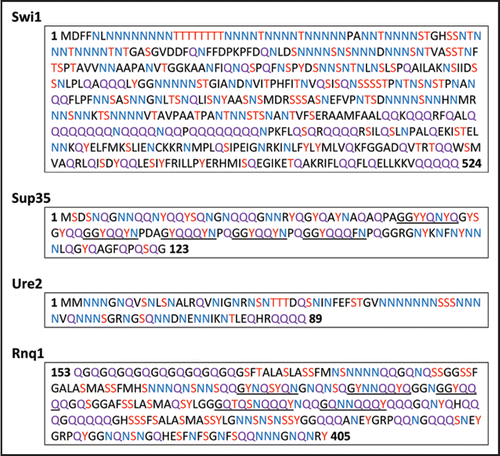
Table 1 Percentage of amino acids present in prion domains of known yeast prions
Table 2 Swi1 is unique among yeast prions proteins due to its nuclear localization and low cellular abundance