Figures & data
Table 1. Efficacy and cellular toxicity screen of 1,4-benzoxazine compounds for PrPSc inhibition in ScN2a cells
Figure 2. 22L-ScN2a cells. Effect of 5,7,8-trimethyl-1,4-benzoxazine derivatives on PrPSc levels by western blot analysis. (A) inhibitors of PrPSc formation. (B) compound TC145 with no inhibitory effects on PrPSc accumulation. Following incubation with each compound, the cells were treated (+) or not treated (-) with PK. All treatments were performed in duplicates (CD1, CD2 for the control, untreated cells and BD1, BD2 for the 5,7,8-trimethyl-1,4-benzoxazines -treated cells). Two representative blots for each active compound, corresponding to different cell passages, are presented in order to show the reproducibility of the acquired results. For PrP-immunostaining, 6H4 was used, whereas for signal normalization, blots were probed with β-actin (C4) antibody.
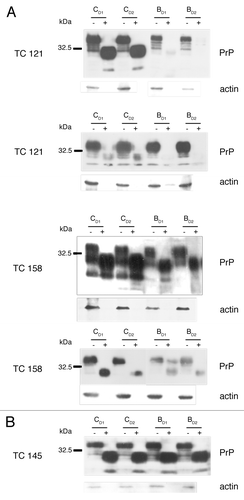
Figure 3. Dose response and toxicity curves. EC50 value, were 50% of the pathological isoform PrPSc is reduced, is 1.17 μM for TC121 (A) and 20 μM for TC158 (C). LD50 value, as derived from the representative toxicity curves, is 10 μM for TC121 (B) and 50 μM for TC158 (D).
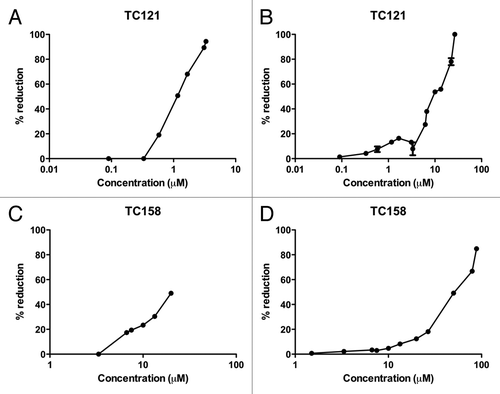
Figure 4. N2a cells. Effect of 5,7,8-trimethyl-1,4-benzoxazine derivatives on PrPC levels by western blot analysis. All treatments were performed in duplicates (CD1, CD2 for the control, untreated cells and BD1, BD2 for the 5,7,8-trimethyl-1,4-benzoxazines-treated cells).
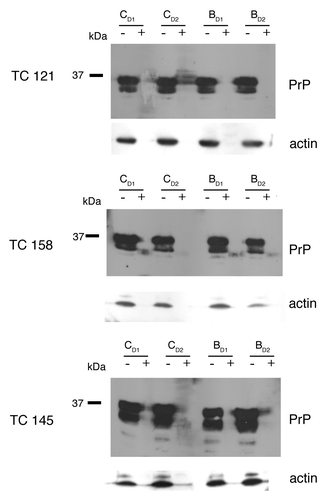
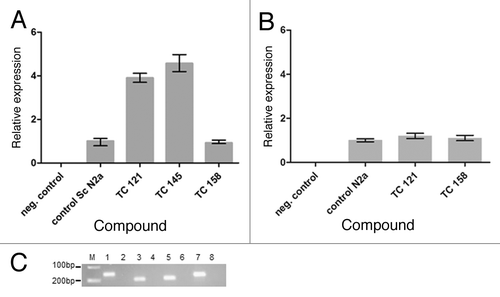
Figure 5. (A) Relative expression of the PRNP gene in untreated “control Sc N2a” cells and cells treated with different 5,7,8,-trimethyl-1,4-benzoxazine derivatives. (B) Relative expression of the PRNP gene in untreated “control N2a” cells and cells treated with TC121 and TC158. As expected, there is no signal in negative control samples without cDNA. (C) 2% agarose gel showing representatively the amplified fragments of the tested genes. M: relative molecular mass markers, 1, 3, 5, 7: PCR products amplified with the β-actin, PrP, RPL32 and GAPDH primers in the presence of cDNA and 2, 4, 6, 8: in the absence of cDNA.