Figures & data
Figure 1. (A) CD spectra of mPrPwt, mPrP-CtoA, S132C, N181C, and S132C/N181C. The proteins were dissolved at a concentration of 0.15 mg/mL in 10 mM NaOAc buffer, pH 7, and their CD spectra immediately recorded. (B) Secondary structure content of each protein determined by CD spectral deconvolution using CDPro software.
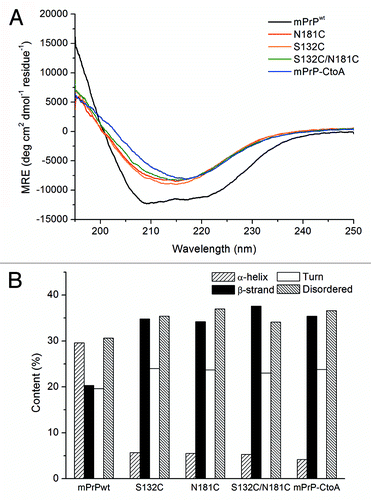
Figure 3. (A and B) CD spectra of S132C/N181C (0.15 mg/mL) in 0.5 mM NaOAc buffer, pH 7, recorded after incubated for the indicated time at 25°C. The protein solution was kept inside the quartz cuvette throughout the experiment. Two structural transitions during the whole time course could be seen. The first transition from an α-helical state to a coil-rich state is shown in (A) and the second transition from a coil-rich state to β-oligomers is shown in (B). The direction of the spectral change is indicated by an arrow. The spectral deconvolution results are shown in (C).
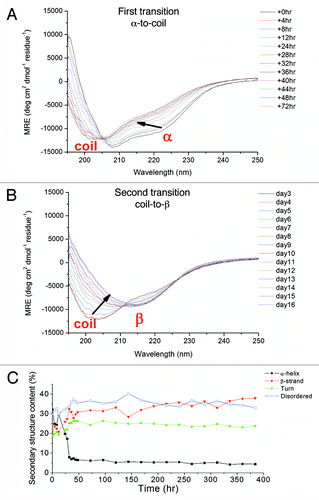
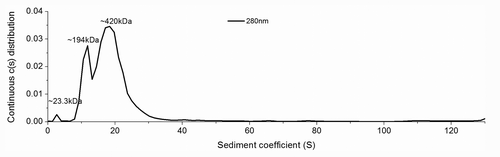
Figure 4. (A) TEM images of S132C/N181C dissolved in 10 mM NaOAc, pH 7. The sample was stained with 2% uranyl acetate and images taken using a Hitachi H-7000 electron microscope at 75 kV. The bars represent 500 nm. (B) ThT fluorescence assay of mPrPwt fibrils (dash line) and S132C/N181C β-PrP oligomers (solid line). S132C/N181C was dissolved at a concentration of 20 μM in 10 mM NaOAc, pH 7, while mPrPwt was incubated at a concentration of 22 μM in 1 M GdnHCl, 3 M urea in PBS at 37°C with shaking at 200 rpm for 5 d. The two protein solutions were mixed with the same volume of ThT working solution and incubated for 1 min, then the fluorescence spectra were recorded from 450 to 600 nm with excitation at 420 nm.
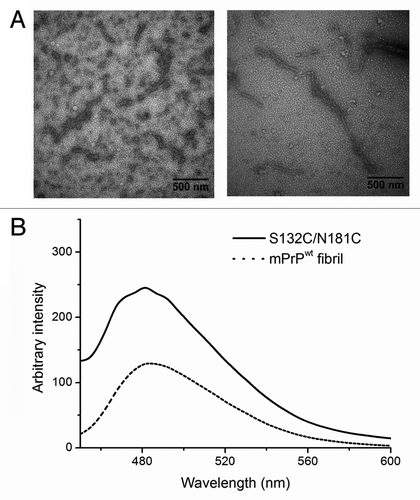
Figure 5. Cytotoxicity of S132C/N181C β-PrP. The β-PrP prepared from S132C/N181C (30 μM) in 10 mM NaOAc, pH 7 was added to the cultured mouse N2a cells. The cytotoxicity was measured by using the MTT assay. The addition of 10 mM NaOAc (pH 7) into the cultured cells was also used as one control. The values are the mean ± standard deviation (n = 6 for S132C/N181C and n = 14 for the others).
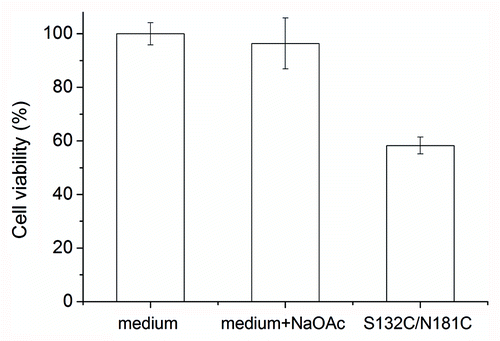
Figure 6. ESR spectra of the spin-labeled mPrP mutants S132R1, N181R1, and S132R1/N181R1. Protein samples were dissolved in 10 mM NaOAc containing 40% sucrose, pH 7.0. The average of the spectra for S132R1 and N181R1 (thin lines) and the spectra of S132R1/N181R1 (bold lines) are shown at the bottom of the figure. ESR spectra were measured either at 275K or 300K.
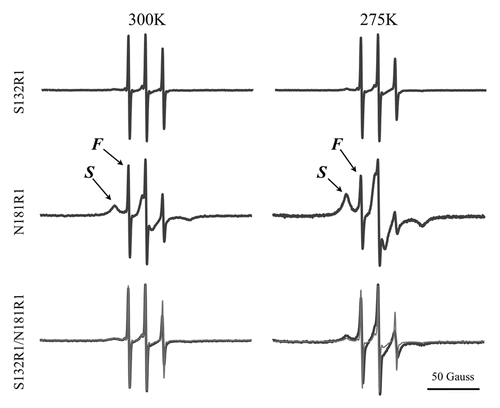
Figure 7. Spontaneous conformational transition of mPrPwt after addition of disulfide-bond reducing agents. (A) The CD spectra of mPrPwt (0.14 mg/mL) in 0.5 mM NaOAc, pH 7 with 1 mM DTT, recorded after incubated for the indicated time at 25°C. The protein was kept inside the quartz cuvette throughout the experiment. The direction of the spectral change is indicated by an arrow. (B) The CD spectrum of mPrPwt (0.14 mg/mL) in 0.5 mM NaOAc, pH 7 with 1 mM TCEP was recorded immediately after sample preparation.
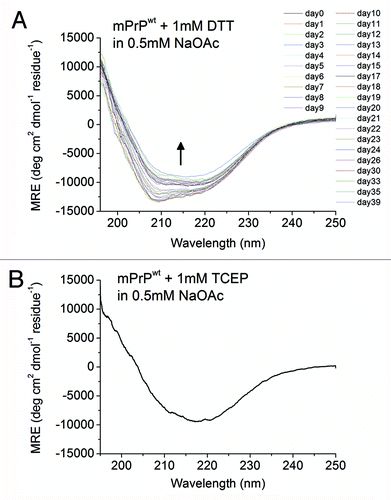
Figure 8. TEM image of mPrPwt dissolved in 0.5 mM NaOAc (pH 7), 1 mM DTT and incubated at 25°C for two days. The sample was stained with 2% uranyl acetate and images taken using a Hitachi H-7000 electron microscope at 75 kV. The bar represents 100 nm.
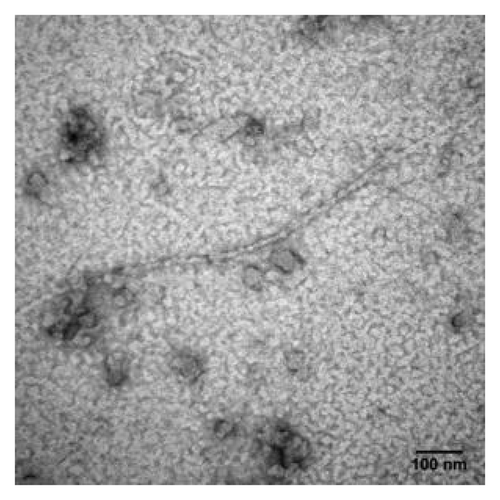
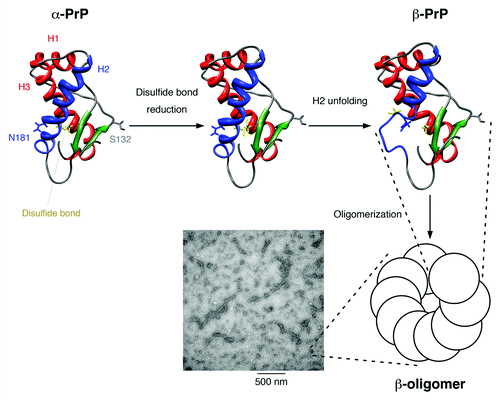
Figure 9. Proposed model for the structural conversion of the mouse prion protein after disulfide bond reduction in neutral condition. The flexible N-terminal domain is not displayed. Helices 1 and 3 are colored red, helix 2 is colored blue, the two β-strands are colored green, and the disulfide bond between Cys-179 and Cys-214 is shown in yellow. The mutation sites used in our study, Ser-132 and Asn-181, are indicated. After the disulfide bond is reduced, helix 2 is no longer stable and is partially unfolded. The disordered part associates and the protein structure is stabilized by the formation of oligomers, which show a characteristic β-sheet signal in the CD spectrum. These β-oligomers have a bead-like morphology under TEM and can associate one-dimensionally into protofibrils.