Figures & data
Figure 1. Design and expression of prion deletion mutants. (A) Schematic representation of murine PrP constructs. The N-terminal basic region is between amino acids 23 and 31. The five octarepeats are between amino acids 51 and 90. The charged cluster upstream of the hydrophobic region is indicated by the box between amino acids 95 and 105. The neurotoxic hydrophobic region is boxed between amino acids 105 and 125. Beta sheets 1 and 2 are labeled S1 and S2, and helices 1, 2 and 3 as H1, H2 and H3 respectively. Balls on sticks represent the glycosylation sites at Asn 180 and Asn 196. The disulphide bridge linking helicies 2 and 3 at Cys 179 and Cys 214 is shown as S-S. (B) SDS-PAGE analysis of all purified prion proteins. Lane 1, PrP 90–231; lane 2, PrP 23–231; lane 3, PrP Δ95–105; lane 4, PrP Δ95–125; lane 5, PrP Δ95–135; lane 6, PrP Δ105–125; lane 7, PrP Δ105–135; lane 8, 5µg of BSA. Numbers to the left of 1B are molecular mass markers and are in kilodaltons (kDa).
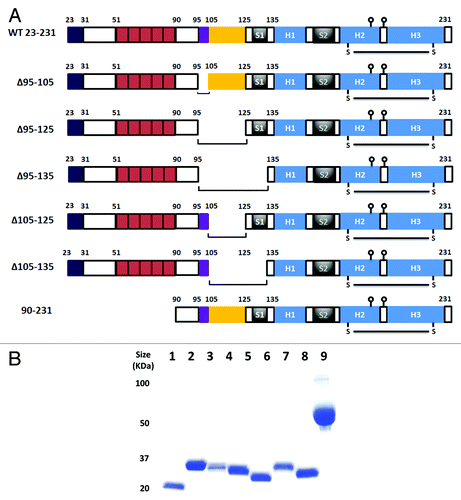
Figure 2. ANS fluorescence. (A) Each prion construct was refolded and assessed for ANS binding in the presence and absence of Cu2+. The concentration of proteins was 35µM, the ANS concentration 600µM and the Cu2+ concentration 50µM. All readings were normalized to control measurement of buffer with ANS alone. Measurements were performed in triplicate and error bars indicate the standard error mean. (B) The effect of pH on ANS fluorescence for WT 23–231 (circle), Δ105–125 (square) and 90–231 (diamond). The filled symbols/solid lines are ANS data in the absence of Cu2+ while the open symbols/dashed lines are ANS data in the presence of Cu2+. The assay conditions were as above with the exception that the pH was independently buffered in each case. Experiments were performed in triplicate and error bars indicate the standard error mean.
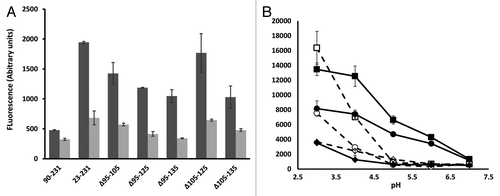
Figure 3. Monoclonal antibody probing of local structure. (A) The epitope locations of each of 5 MAbs used in the study drawn on a schematic of the prion protein structure, the N-terminus is omitted for clarity. (B) Comparative ELISA of WT 23–231 and Δ105–125 with each of the MAbs shown. Each protein antigen was normalized to 5µM before coating to the plate.
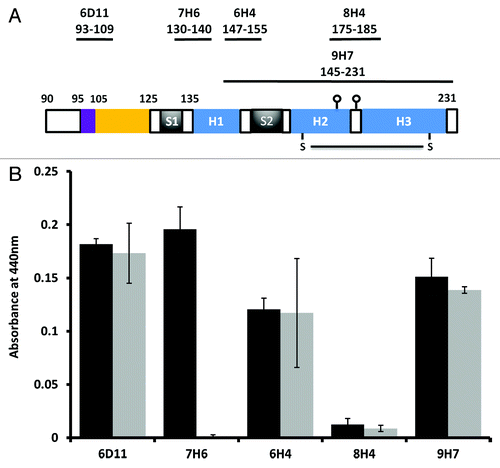
Table 1.CD spectroscopy of purified prion proteins after analysis of α helix and β sheet content using the K2D3 algorithm.
Figure 4. Expression of prion deletions in Sf9 cells as fusion proteins with GFP. (A) Western blot of Sf9 cells infected with each prion-GFP fusion with mAb 6H4 at two days post infection. (B) Mean fluorescence of each prion-GFP fusion at 2 d post infection normalized to the 6H4 signal. (C) Fluorescence microscopy of Sf9 cells expressing GFP tagged prion protein. The upper panel is WT 23–231-GFP and the lower is Δ105–125-GFP. The scale bars indicate 400µm.
Figure 5. Binding to synthetic lipids. ELISA data for binding of 6H4 to each fraction of a sucrose gradient following flotation assay. Purified prion proteins WT 23–131, 90–231 and Δ105–125 were incubated with POPG (solid lines) or PC (dashed lines) vesicles as described, prior to underloading the gradient. The samples were: WT 23–231 with no lipid (◆), WT 23–231 with PC (■), WT 23–231 with POPG (▲), Δ105–125 with PC (X), Δ105–125 with POPG (□) and 90–231 with POPG (●). 93–231 with PC was not done. Fraction 1 represents the top and fraction 8 the bottom of the gradient. ELISA measurements were performed in triplicate and error bars indicate the standard error mean.
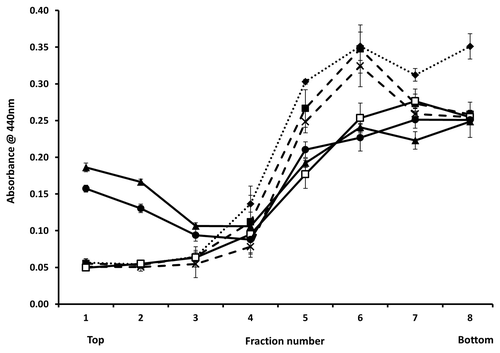
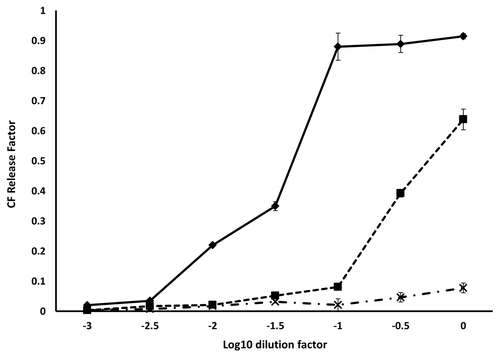
Figure 6. Fluorescence dequenching of CF loaded vesicles by prion protein binding. The initial concentration of each protein was 10 µM. The concentration of lipid remained constant over the dilution series at 250 µg/ml. The samples were: WT 23–231 (◆), 90–231 (■) and Δ105–125 (X). Measurements were performed in triplicate and error bars indicate the standard error mean.