Figures & data
Figure 1. Affinity of V-D to PrP, as measured using the Biacore system. (A) The interaction between PrPc (90–231) and V-D2 showed high binding. (B) The interaction between PrPc (90–231) and V-D3 showed no binding affinity. (C) The interaction between PrPc (90–231) and V-D2, after saturating with the3F4 mAb. (D) The interaction between PrPc (101–130) and V-D2, after saturating with the 3F4 mAb. Experiments were performed at least 3 times and similar results were obtained. The figures express the representative results of the experiments.
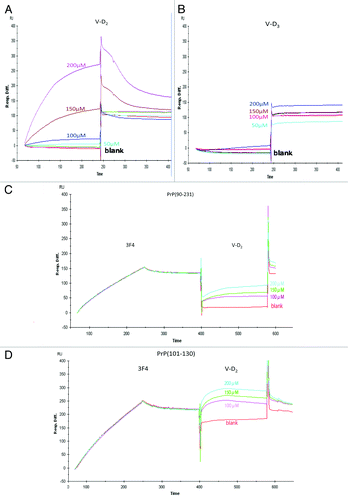
Table 1. Binding kinetics of V-D2 and V-D3 to Hu-rPrP(90–231)
Figure 2. Reactivity of mAbs against PrPc with V-D2 by ELISA. (A) The 3F4 epitope on PrPc was affected by V-D2 in a dose-dependent manner. The blue line indicates signals for Hu-rPrPc (90–231), and the red line indicates signals for PrPc (101–130). (B) The SAF70 epitope on PrPc was not affected by V-D2. The blue line indicates signals for Hu-rPrPc (90–231), and the green line indicates signals for PrPc (141–170). Each experiment was performed 3 times by triplicates, and the values express means ± SD *p < 0.006,**p < 0.003 vs. PrPc(101–130) without V-D2, †p < 0.005, ††p < 0.01, †††p < 0.05 vs. Hu-rPrPc(90–231) without V-D2.
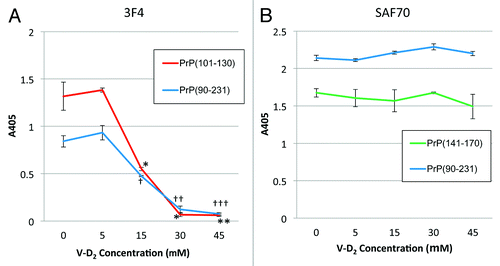
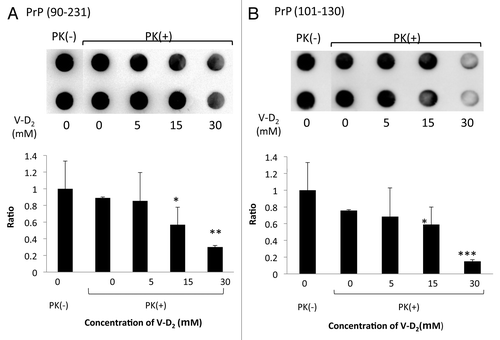
Figure 3. Sensitivity of PrPc to protease K following incubation with V-D2. V-D2 increased the sensitivity of Hu-rPrPc (90–231) (A) and PrPc (101–130) (B) to PK in a dose-dependent manner. The average pixel density of each spot was measured by NIH image analysis after subtracting the mean background pixel density from that of the test spots. Experiments were performed 3 times by duplicates, and the representative results were expressed. Values are means ± SD of three experiments. *p < 0.05, **p < 0.02, ***p < 0.01 vs. Hu-rPrPc (90–231) or PrPc (101–130) with PK in the absence of V-D2.