Figures & data
Figure 1.hsp104-G254D and hsp104-G730D strains show inefficient [PSI+] propagation. (A) hsp104-G254D and hsp104-G730D cells display a sectoring [PSI+] phenotype as a result of inefficient [PSI+] inheritance. (B) Lysates of sectoring hsp104-G254D and hsp104-G730D cells with strong [PSI+] cells ([PSI+]) and [psi−] cells ([psi−]) as controls were analyzed by SDD-AGE analysis followed by western blot and blotting for Sup35. These results were reproduced at least three times. An example of the shift is shown. The general loss of the lower aggregate species and the decrease in aggregated Sup35 is reproducible. The appearance of monomeric Sup35 on SDD-AGE western blots is more variable, even with controls, for unknown reasons.
![Figure 1.hsp104-G254D and hsp104-G730D strains show inefficient [PSI+] propagation. (A) hsp104-G254D and hsp104-G730D cells display a sectoring [PSI+] phenotype as a result of inefficient [PSI+] inheritance. (B) Lysates of sectoring hsp104-G254D and hsp104-G730D cells with strong [PSI+] cells ([PSI+]) and [psi−] cells ([psi−]) as controls were analyzed by SDD-AGE analysis followed by western blot and blotting for Sup35. These results were reproduced at least three times. An example of the shift is shown. The general loss of the lower aggregate species and the decrease in aggregated Sup35 is reproducible. The appearance of monomeric Sup35 on SDD-AGE western blots is more variable, even with controls, for unknown reasons.](/cms/asset/6eaf19e0-9381-4899-baf3-8919b770e771/kprn_a_10926547_f0001.gif)
Figure 2.hsp104-G254D and hsp104-G730D cells propagate the original strong [PSI+] variant. (A) hsp104-G254D and hsp104-G730D cells (G254D and G730D, respectively) and diploids from the mating of hsp104-G254D and hsp104-G730D to wild type [psi−] cells (WT/G254D and WT/G730D) were spotted on rich media (YPD) and media lacking adenine (SD-Ade). Strong [PSI+] and [psi−] cells are spotted for comparison. The second spot in each row is a 5-fold dilution of the first spot. (B) Full tetrads from the sporulation of the diploids in (A) were spotted for each mutant as indicated. Each haploid progeny in the tetrad is labeled, A–D. Strong [PSI+] and [psi−] are spotted for color comparison. The second spot in each row is a 5-fold dilution of the first spot. (C) Representative tetrads from (B) were analyzed by SDD-AGE analysis and western blot. The letters A–D correspond to the haploids with the same label in (B) and weak [PSI+] (Weak), strong [PSI+] (Strong) and [psi−] were analyzed for comparison. SDD-AGE analysis was performed four times with haploids from both hsp104-G254D and hsp104-G730D cells.
![Figure 2.hsp104-G254D and hsp104-G730D cells propagate the original strong [PSI+] variant. (A) hsp104-G254D and hsp104-G730D cells (G254D and G730D, respectively) and diploids from the mating of hsp104-G254D and hsp104-G730D to wild type [psi−] cells (WT/G254D and WT/G730D) were spotted on rich media (YPD) and media lacking adenine (SD-Ade). Strong [PSI+] and [psi−] cells are spotted for comparison. The second spot in each row is a 5-fold dilution of the first spot. (B) Full tetrads from the sporulation of the diploids in (A) were spotted for each mutant as indicated. Each haploid progeny in the tetrad is labeled, A–D. Strong [PSI+] and [psi−] are spotted for color comparison. The second spot in each row is a 5-fold dilution of the first spot. (C) Representative tetrads from (B) were analyzed by SDD-AGE analysis and western blot. The letters A–D correspond to the haploids with the same label in (B) and weak [PSI+] (Weak), strong [PSI+] (Strong) and [psi−] were analyzed for comparison. SDD-AGE analysis was performed four times with haploids from both hsp104-G254D and hsp104-G730D cells.](/cms/asset/d75c36e9-9b9e-4cf9-9ed4-5fdfd836eb22/kprn_a_10926547_f0002.gif)
Table 1. Hsp104 mutants display defects in ATP hydrolysis under physiological salt conditions
Figure 3. Hsp104-G254D and Hsp104-G730D mutant proteins form hexamers in vitro. Hsp104 (blue), Hsp104-G254D (green), and Hsp104-G730D (red) were incubated with (solid color) or without (hatched) 5 mM ATP for 10 min then subjected to ultracentrifugation through a linear (10–35%) glycerol gradient. Equal volume fractions were collected and the amount of Hsp104 protein in each fraction was analyzed by SDS-PAGE and western blot. Individual bands were quantified and the amount of Hsp104 in each fraction was plotted as a percent of the total Hsp104 protein. The graph shows the data from one assay. This assay was performed three times and all gave similar results.
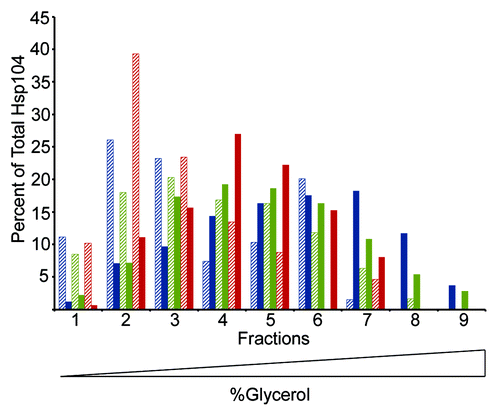
Figure 4.hsp104-G254D and hsp104-G730D display different levels of non-prion disaggregation. The thermotolerance of HSP104, hsp104Δ, hsp104-G254D, and hsp104-G730D cells was tested. Cells were first grown at 37 °C in liquid culture to induce HSP104 expression, heat-shocked at 50 °C for 10 to 30 min as indicated, and then spotted on rich media. Control cells (No Heat) were plated without heat shock. Thermotolerance assays were repeated four times and all showed similar results.
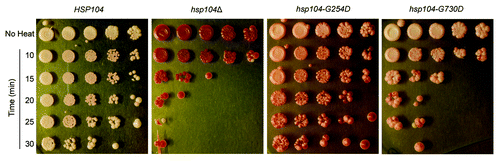
Figure 5. Neither hsp104-G254D nor hsp104-G730D can propagate weak [PSI+]. (A) [psi-] hsp104-G254D and [psi-] hsp104-G730D cells were mated to either weak [PSI+] or the weak [PSI+] variant Sc37. Heterozygous HSP104 and hsp104 mutant diploids (Weak/mut and Sc37/mut) from these matings and haploid parents (Cured, Weak, Sc37) were spotted on YPD to assess phenotype. The second spot in each row is a 5-fold dilution of the first spot. (B) Diploids of the mutants crossed to weak [PSI+] (Weak/Mut in A) were sporulated and 16 tetrads dissected. Two representative tetrads on YPD are shown. Each haploid is labeled A–D. (C) Diploids of weak [PSI+] crossed to the mutants (Weak/Mut in A) and a representative tetrad from these heterozygous diploids were subjected to SDD-AGE and western blot analysis to determine Sup35 aggregate distribution. SDD-AGE analysis was performed twice using four distinct tetrads.
![Figure 5. Neither hsp104-G254D nor hsp104-G730D can propagate weak [PSI+]. (A) [psi-] hsp104-G254D and [psi-] hsp104-G730D cells were mated to either weak [PSI+] or the weak [PSI+] variant Sc37. Heterozygous HSP104 and hsp104 mutant diploids (Weak/mut and Sc37/mut) from these matings and haploid parents (Cured, Weak, Sc37) were spotted on YPD to assess phenotype. The second spot in each row is a 5-fold dilution of the first spot. (B) Diploids of the mutants crossed to weak [PSI+] (Weak/Mut in A) were sporulated and 16 tetrads dissected. Two representative tetrads on YPD are shown. Each haploid is labeled A–D. (C) Diploids of weak [PSI+] crossed to the mutants (Weak/Mut in A) and a representative tetrad from these heterozygous diploids were subjected to SDD-AGE and western blot analysis to determine Sup35 aggregate distribution. SDD-AGE analysis was performed twice using four distinct tetrads.](/cms/asset/816499c2-9f78-4531-84b1-17be2085dfc5/kprn_a_10926547_f0005.gif)
Figure 6.hsp104-G730D can propagate multi-dot high [RNQ+]. hsp104-G730D cells were mated to s.d. low (low), medium (med), high (high), very high (v.high) and m.d. high (m.d. high) [RNQ+] and then sporulated. The hsp104-G730D haploid progeny from four separate tetrads of each variant mating, along with the unmated [RNQ+] variants as controls (WT), were analyzed by SDD-AGE and western blot.
![Figure 6.hsp104-G730D can propagate multi-dot high [RNQ+]. hsp104-G730D cells were mated to s.d. low (low), medium (med), high (high), very high (v.high) and m.d. high (m.d. high) [RNQ+] and then sporulated. The hsp104-G730D haploid progeny from four separate tetrads of each variant mating, along with the unmated [RNQ+] variants as controls (WT), were analyzed by SDD-AGE and western blot.](/cms/asset/2edaf5eb-30f3-4bfe-878f-b87d52302535/kprn_a_10926547_f0006.gif)