Figures & data
Figure 1. PrP sequence emphasizing segments relevant to this paper. The sequence elements shown are nearly identical in Hu, SHa and MH2M PrP’s, sharing all of the charged residues shown. MH2M, a mouse-hamster chimera, lacks Gly55. Hu numbering is used. The TM1 segment (112–134) and terminal signal sequences are shown as gray boxes. The mutations shown include KH-II (K110I, H111I) and A3V (A113V, A115V, A117V), principally affecting TM1 hydrophobicity, and P102L, all studied in mice.Citation37 The R136D, K106E and K110E mutations alter ΔCh across TM1 and the TSE25 (K23T, K24S, R25E) mutation at the mature N-terminus alters ΔCh across the signal peptide.
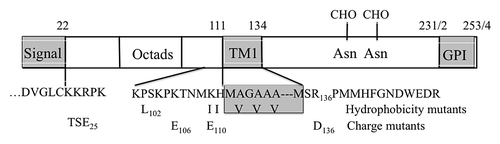
Figure 2. PrP fusions expressed in yeast. Left; models of PrP fusions spanning the membrane. From the top, an HAh6 Ctm fusion, an HAh6 Ntm fusion, an HAh9βla Ctm fusion and an HAh6αβla Ctm fusion. Ntm forms of the latter species would also be inverted about the TM1 segment. The unstructured mature N-terminal segment is shown as a loop, TM1 as a four-loop helix and the globular C-terminal segment as an ovoid. In both Ctm and secreted forms this segment may be glycosylated at two sites, indicated by CHO. In Ntm orientation the C-terminal segments are retained in the cytoplasm and not glycosylated. HA is the triple HA tag, h6 and h9 are His6 and His9 Ni-binding tags, βla is the mature sequence of β-lactamase, as normally secreted, and α is a 30 residue fragment of pre-pro-α factor that includes two additional N-glycosylation sites and a site for cleavage by ER-Kex2p. Cleavage releases βlactamase in the ER lumen for secretion. Right; predicted sizes in kDa are shown, assuming that, in Ctm and secreted forms, the secretion signal is removed and all sites receive core N-glycosylation. Protease K treatment of intact microsomes should remove the HA tags from Ntm forms, should cleave about 8 kDa from the N-terminus of Ctm forms but should not affect secreted forms. Endoglycosidase H treatment of ruptured microsomes should remove about 2.3 kDa for each core structure cleaved. Fragment sizes for the Ctm HAh6αβla fusions are shown after removal of 29 kDa βlactamase by Kex2p cleavage.
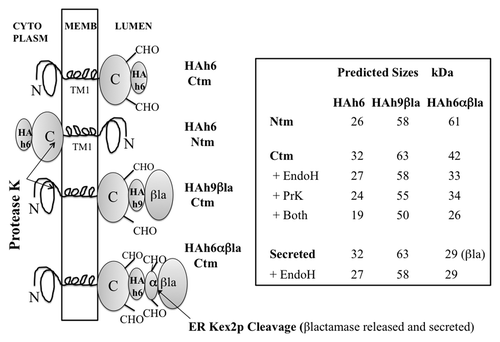
Figure 3. In vitro expression of PrP-HAh6 fusions. T7 run-off transcripts were translated by rabbit reticulocyte extracts in the presence of dog pancreas microsomes in the presence or absence of NYT, a competitive inhibitor of glycosylation (lanes 1–2, 5–6, 8–9). Protease K treatment was performed in the presence (lane 4) or absence (lanes 3, 7, 10) of 1% triton × 100.
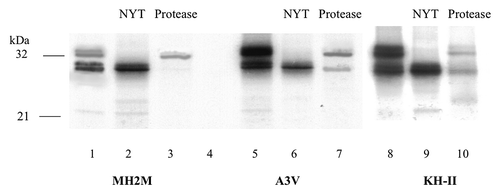
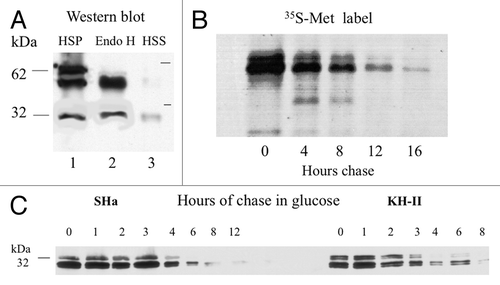
Figure 4. (A) Yeast expression of SHa-HAh6 and its KH-II mutant. Spheroplasts were isolated from exponential phase cells and separated at 3 000 g into low-speed pellet (LSP) and supernatant (LSS) fractions. The LSS was then separated at 250 000 g into high-speed pellet (HSP, membranes) and supernatant (HSS, cytoplasm) fractions. The lysed HSP sample was hydrolyzed with endoglycosidase H (Endo H). Samples representing constant cell number were fractionated by SDS-PAGE and detected by western blot using anti-HA monoclonal antibody. (B) Membrane localization of MH2M- and A3V-HAh6 fusions. Samples were fractionated by SDS-PAGE and detected as in A. Spheroplasts were isolated from exponential phase cells and separated at 3000 g into low-speed supernatant (lanes 1, LSS) and pellet (lanes 2, LSP) fractions. Spheroplasts in the LSS were lysed and hydrolyzed with endoglycosidase H (Endo H, lanes 3). The LSS was separated at 250 000 g into supernatant (HSS, cytoplasm) and pellet (HSP, membranes) in buffer A1 (lanes 8–9), with the addition of 1% triton × 100, 0.1% sarkosyl (lanes 4–5) or in 0.1M pH 11.5 carbonate buffer (lanes 6–7). Lanes 10 show concentrated culture supernatant. Sample loads in lanes 6–10 were 5-fold higher than in lanes 1–5.
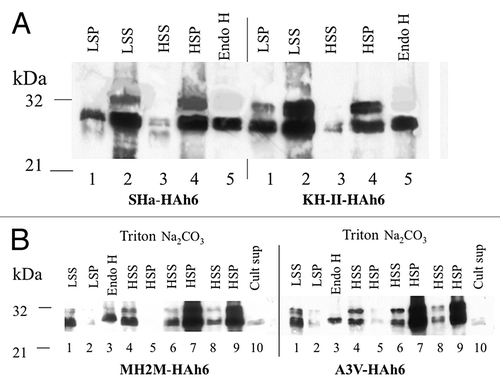
Figure 5. (A) Yeast expression of A3V-HAh9βla. Cells were hydrolyzed with zymolyase and the low-speed supernatant spheroplast fraction separated at 250 000 g into pellet (HSP, lane 1) and supernatant (HSS, lane 3). Fractions were analyzed by SDS-PAGE and detected using monoclonal anti-β-lactamase serum. Lane 2 shows the products of endoglycosidase H treatment of the HSP fraction in lane 1. (B) pulse-chase half-life analysis. Cells in a parallel culture were labeled for 20 min with 35S methionine. After washing and chase with cold methionine for the indicated times, proteins immunoprecipitated with anti-β-lactamase serum were detected by autoradiography. (C) half-lives of SHa- and KH-II-HAh6 fusions determined by western blot. Exponential phase cells expressing SHa- or KH-II-HAh6 fusions from the GAL1 promoter were shifted from galactose to glucose medium at time zero. Samples were removed at the indicated times for western blot analysis using anti-HA monoclonal antibody. Band densities were determined by densitometry.
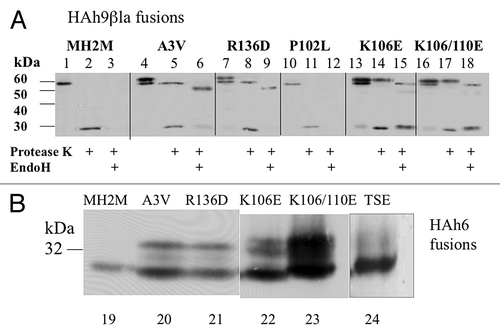
Table 1. % N-glycosylated (Ctm + secreted) PrP fusion species
Figure 6. Effects of A3V, R136D, P102L, K106D, K110D and TSE25 mutations on translocation of MH2M fusions. (A) Cells expressing the indicated mutants of MH2M-HAh9βla were converted to spheroplasts, gently lysed, and microsomal membranes (3000 g supernatants, 250 000 g pellets) were isolated. Proteins were detected by western blot, using anti-β-lactamase serum after no treatment (lanes 1, 4, 7, 10, 13, 16), after exposure to protease K (lanes 2, 5, 8, 11, 14, 17), or after subsequent hydrolysis with endoglycosidase H (lanes 3, 6, 9, 12, 15, 18). β-lactamase released by protease K is detected as a 29 kDa species. (B) Cells expressing the indicated mutants of MH2M-HAh6 were broken with glass beads and proteins in the low speed supernatant were detected by western blot using anti-HA monoclonal antibody. Lanes 19–23 are all derived from the same blot. Lane 24 is from a separate blot run under identical conditions.