Figures & data
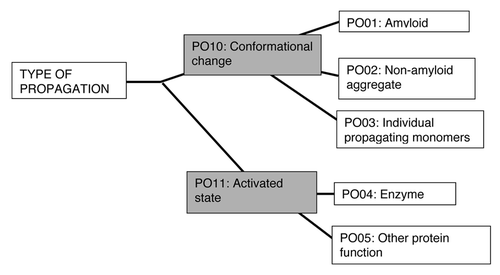
Figure 1. Classification of prion/prion-like phenomena into broad groups. The prions and quasi-prions can be apportioned into four different broad groups, plus the transcellular prionoids. A narrower prion definition would place Group II in the Quasi-Prions, or conversely, a broader definition would place Group III in the Prions. Group I corresponds to the Type Am* prions in the current PrionHome database release (http://libaio.biol.mcgill.ca/prion), Group II to the Type Am and Ac prions, Group III to the Quasi-prions from Alberti, et al.Citation4 and Group IV to the remaining Quasi-prions. In each box, the protein names and PrionHome database accessions are listed for examples from each category (for prions the name of the prion phenomenon is also given). Complete sets of the categories are available in the database. The database also includes: interactors of prion proteins, paralogs and orthologs of prionogenic proteins, and candidate prion sequences predicted algorithmically,Citation3-Citation5 and tools for visualizing alignments and structures for database entries. Specific experimental details about each prion/prion-like phenomenon are listed in the comments sections of individual database entries.Citation3
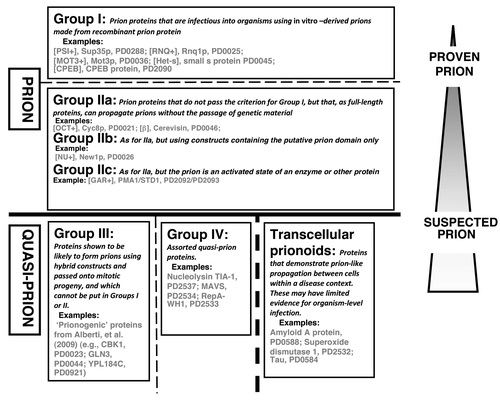
Figure 2. An example of a prion ontology hierarchical tree. The “type” of propagation can be classified using a simple hierarchical tree of terms. The most specific terms arise at the leaf nodes of the tree, with more general terms at appropriate internal nodes. Proteins underlying prion or prion-like phenomena can be annotated with terms from multiple such trees. According to this tree, for example, the yeast protein Rnq1p would be annotated with the terms PO01 and PO10.