Figures & data
Figure 1. Average D/N ratios for all samples. (A) D/N ratios for sCJD samples. C (black); FL (dark-gray); OL (gray); OB (light-gray); SC (white). (B) D/N ratios for non-CJD samples. Dark-gray columns represent NC (13–19) and light-gray columns represent NNC (20–30). The data was obtained from PrP226* assay. Sequential numbers of samples are as in . C, cerebellum; FL, frontal lobe; OL, occipital lobe; OB, olfactory bulb; SC, spinal cord.
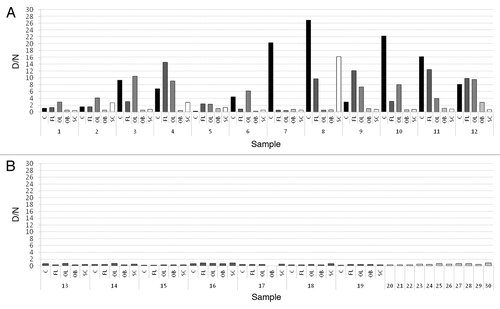
Figure 2. (A) Percentage of samples from certain brain region of 12 sCJD cases, which were defined as positive. (B) Results of PrP226* assay for all sCJD cases. Sequential numbers of samples are as in
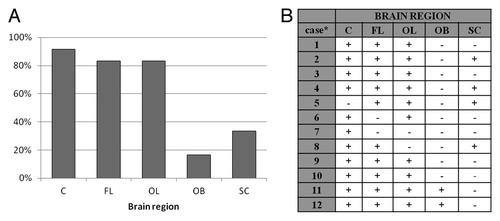
Figure 3. Denaturation profiles. D/N ratios are ploted as the function of the GdnSCN concentration. Sequential numbers of samples are as in . C, cerebellum; FL, frontal lobe; SC, spinal cord.
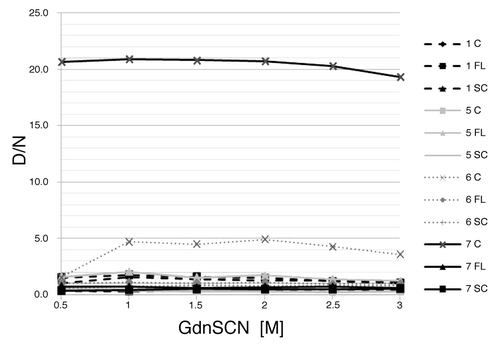
Figure 4. Comparison of A450 values of non-denatured samples (all brain regions included). The first bar represents the average value of non-denatured non-CJD samples (NC, NNC). sCJD samples were divided in two groups – samples, that were defined as negative (D/n < 1.2) were included in group 1, samples, that were defined as positive (D/n > 1) were included in group 2. The second bar represents the average value of non-denatured sCJD samples from group 1 and the third bar represents the average value of non-denatured sCJD samples from group 2.
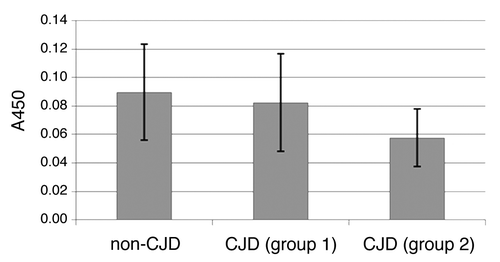
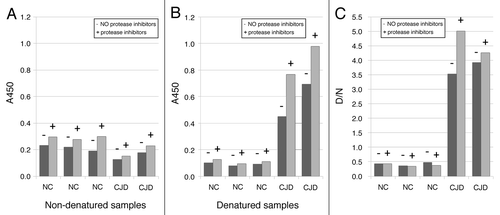
Figure 5. Influence of protease inhibitors in homogenization buffer on the amount of PrP226*. Samples were homogenized in buffer with (+) or without (-) of protease inhibitors. A450 values are shown for non-denatured (A) and denatured (B) samples.