Figures & data
Figure 1. Detailed atomic resolution structures of amyloid leads to different physical properties, which in turn produce varied biological functions. (A–C) Structural models depicting the amyloid cross-β molecular architecture. (A) the crystal structure of GNNQQNY (2OMM.pdb),Citation7 (B) a model of fibrous nanocrystals formed by KFFEAAAKKFFE,Citation8 (C) a model structure of fibrils formed by VIYKI.Citation9 (D–H) schematic illustrations of morphologies that may result in different biological properties. (D) Illustrations of flexible and heterogeneous fibrils compared with (E) rigid and crystal-like fibrils. (F) Illustrations showing long and thermally, mechanically and/or enzymatically stable fibrils compared with (G) short, fragile, and unstable fibrils with non-interacting surfaces and (H) fibrils with interacting or sticky surfaces.
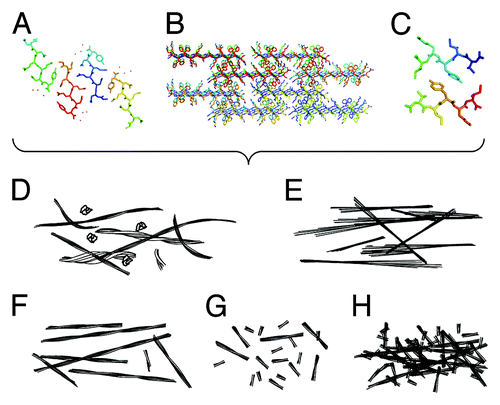
Figure 2. Structure and morphology of different amyloid aggregates assembled from KFFEAAAKKFFE Citation8 and variants Citation24 results in altered morphology and assembly properties. Images show aggregated peptide samples 1: KFFEAAAKKFFE in PBS, 2: RFFEAAAKRFFE in PBS, 3: AFFEAAAAKFFE in water, and 4: KFFEAAAAKFFE in water. Upper row shows negative stain transmission electron microscope images (black scale bars = 200 nm). Lower row shows atomic force microscopy height images (white scale bars = 2 μm).
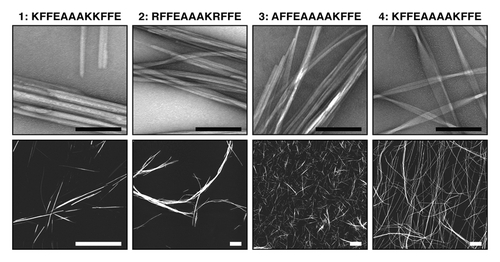
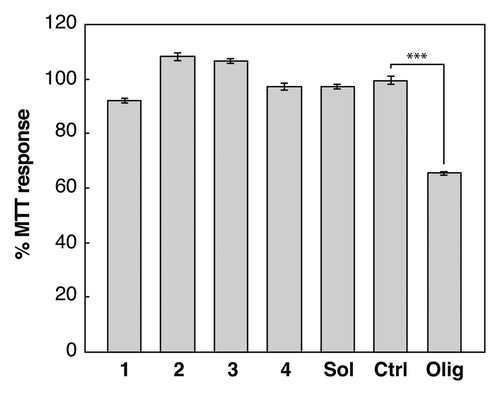
Figure 3. The effect of assembled peptide morphology and composition on neuroblastoma cells. MTT assays (Molecular Probes) were performed according to the manufacturers protocol. A neuroblastoma cell line (SH-SY5Y) was treated with 10 μM monomer equivalent concentrations of aggregated peptide samples 1: KFFEAAAKKFFE in PBS 2: RFFEAAAKRFFE in PBS, 3: AFFEAAAAKFFE in water, and 4: KFFEAAAAKFFE in water (). Cells were also treated with soluble KFFEAAAKKFFE in water (labeled Sol), a buffer only control (labeled Ctrl) or Aβ42 oligomers (labeled Olig). Changes in the MTT response after 24 h were monitored and are shown relative to the buffer control and error bars correspond to SEM over 3 replicates. Only Aβ42 oligomersCitation16,Citation26 induce cellular dysfunction resulting in a reduction to 60% compared with control. The student t test comparison between olig and ctrl showed significance ofP < 0.001 labeled ***.