Figures & data
Figure 1. Region of the 15N-HSQC NMR spectra for the ShPrP90–232 before (A) and after (B) the addition of an equimolar amount of LPS.
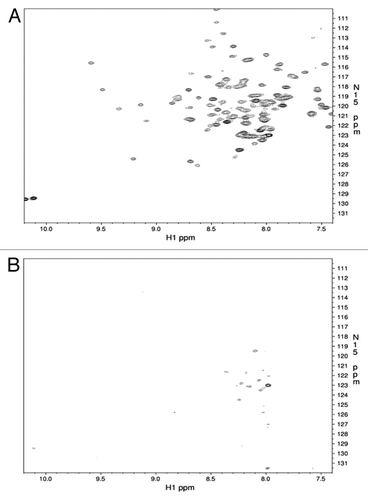
Figure 2. Electron microscope images of the LPS mediated conversion of the ShPrP (90–232) protein. (A) LPS micelles at 0.5 mg/mL in water, pH 7.0 (magnification 71 000×, scale bar 50 nm). (B) The same LPS “micelles” shown in (A) after addition of 0.5mg/mL ShPrP (magnification 56 000×, scale bar 200 nm). (C and D) The sample after the addition of 150 mM NaCl, 0.1% NaN3 and incubated at 37 °C for 48 h magnification 110 000× and 14 000×, scale bars: 50 nm and 0.5 um respectively). Inset in was collected at 110 000× (scale bar 200 nm). Small fibrils can be seen in C. Samples were stained with lead acetate (A) or uranyl acetate (B–D).
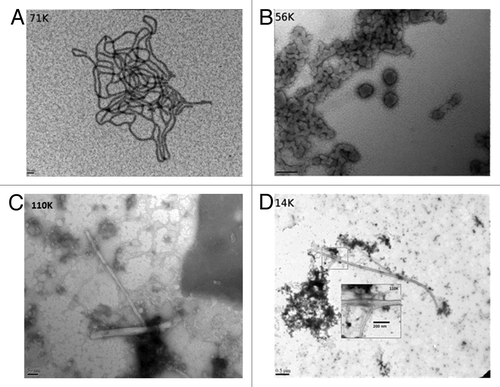
Figure 3. (A) SDS-PAGE gel of proteinase-K treated ShPrP isoform.Lane 1: LPS-generated ShPrPβ. Lane 2: ShPrP (90–232). Lane 3: ShPrP digested with PK (PK:ShPrP 1:1000). Lane 4: Molecular weight ladder (molecular weights on the right). Lane 5: PK:ShPrPβ 1:1000. Lane 6: PK:ShPrPβ 1:750. Lane 7: PK:ShPrPβ 1:500. Lane 8: PK:ShPrPβ 1:250. (B) CD spectra for the ShPrP (90–232) with 1:1 w/w ratio of LPS at 0 incubation time.
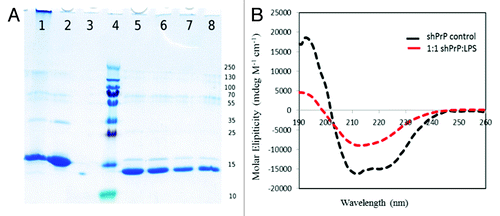
Figure 4. CD spectra for 25 μM of ShPrP (90–232) with various concentration (w/w ratios) of LPS (1:6 300 μM LPS, 1: 3–150 μM LPS, 1:1.5–75 μM LPS, 1:0.75–37.5 μM LPS, 1:0.38–18.5 μM LPS, 1:0.17–9.5 μM LPS, 1: 0.09–5 μM LPS, and 1:0.01–1 μM LPS). Spectra were collected immediately after adding LPS, except for the 1:0.01 w/w ratio of LPS with shPrP(90–232), which was incubated an additional 4 wk to ascertain any changes in secondary structure.
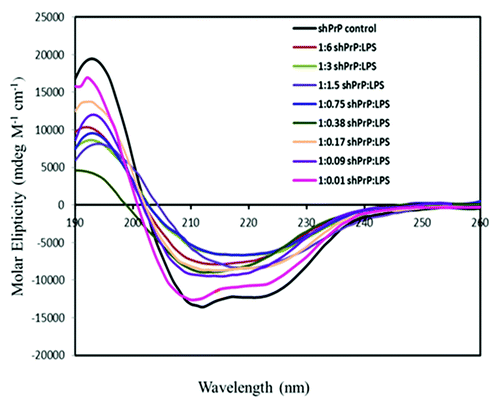
Table 1. Secondary structure content (measured by CD) of LPS-converted prions
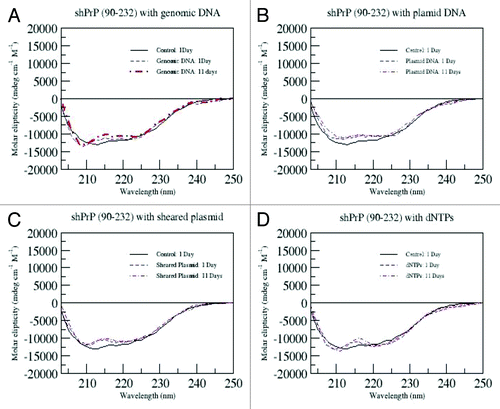
Figure 5. CD spectra for the ShPrP (90–232) with various molar ratios of DOPE, SDS, POPG, and urea at 0 incubation time. (A) CD spectra of the prion propagation studies using DOPE (diluted from 1:64 to 1:16 PrPC:DOPE (w/w)). (B) Urea induced oligomers and fibrils (for comparison of secondary structure content). (C) SDS (1:0.2 to 1:0.0125 PrPC:SDS). D. POPG (diluted from 1:0.5 to 1:0.065 of ShPrP [90–232]:POPG). Each diluted sample was incubated an additional 19 d to ascertain any changes in secondary structure.
![Figure 5. CD spectra for the ShPrP (90–232) with various molar ratios of DOPE, SDS, POPG, and urea at 0 incubation time. (A) CD spectra of the prion propagation studies using DOPE (diluted from 1:64 to 1:16 PrPC:DOPE (w/w)). (B) Urea induced oligomers and fibrils (for comparison of secondary structure content). (C) SDS (1:0.2 to 1:0.0125 PrPC:SDS). D. POPG (diluted from 1:0.5 to 1:0.065 of ShPrP [90–232]:POPG). Each diluted sample was incubated an additional 19 d to ascertain any changes in secondary structure.](/cms/asset/941c5b2a-f225-4a74-98eb-4cf229092a26/kprn_a_10928939_f0005.gif)
Table 2. Secondary structure content of urea, DOPE, POPG, and SDS mediated conversion of prion protein
Figure 6. Resolution enhanced native acidic gel electrophoresis (RENAGE) shows different size and stability of oligomers formed from the different conversion methods. Conversion of recombinant ShPrP (90–232) is compared for urea/salt conversion (3M urea 20 mM sodium acetate pH 4 and 200 mM NaCl), POPG conversion (0.05:1 POPG to PrP [w/w], incubated at 37 °C for 4 d), SDS conversion (0.02% SDS), and LPS conversion (1:0.09 PrP to LPS, incubated at 37 °C for 2 d), 8 μg of each sample is loaded. The latter is from MoPrP (90–231) cross-linked non-specifically using PICUP.
![Figure 6. Resolution enhanced native acidic gel electrophoresis (RENAGE) shows different size and stability of oligomers formed from the different conversion methods. Conversion of recombinant ShPrP (90–232) is compared for urea/salt conversion (3M urea 20 mM sodium acetate pH 4 and 200 mM NaCl), POPG conversion (0.05:1 POPG to PrP [w/w], incubated at 37 °C for 4 d), SDS conversion (0.02% SDS), and LPS conversion (1:0.09 PrP to LPS, incubated at 37 °C for 2 d), 8 μg of each sample is loaded. The latter is from MoPrP (90–231) cross-linked non-specifically using PICUP.](/cms/asset/e7614a27-003c-4864-aa44-3c265784b992/kprn_a_10928939_f0006.gif)