Figures & data
Figure 1. Chemical structures of thiophene based amyloid ligands and fluorescence images of stained PrP deposits in brain tissue sections from mice infected with mSS or mCWD. (A) PTAA, a polydisperse polythiophene. (B) p-FTAA, a chemically defined pentameric oligothiophene. (C) p-KTAA, a chemically defined pentameric oligothiophene. (D) h-FTAA, a chemically defined heptameric oligothiophene. Scale bar represents 50 μm.
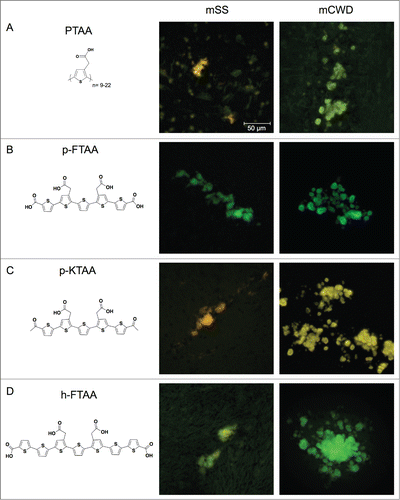
Figure 2. Emission and excitation spectra of thiophene based amyloid ligands bound to mSS (dotted line) or mCWD (black line) deposits. Emission spectra for (A) PTAA, (B) p-FTAA, (C) p-KTAA, and (D) h-FTAA. The emission spectra were recorded using excitation at 488 nm. Excitation spectra for (E) PTAA, (F) p-FTAA, (G) p-KTAA, and (H) h-FTAA. The excitation spectra were recorded having the emission locked for each probe and prion strain within the respective emission window.
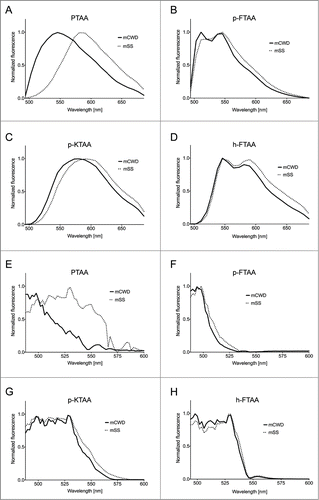
Table 1. Mean Euclidean distances for the emission and excitation profiles within and between the prion strains
Figure 3. Heat maps and dendrograms for Euclidean distances between pairs of emission spectra from 50 different regions from PrP deposits associated with the respective prion strain. Rows and columns in the heat maps are ordered according to hierarchical clustering of the spectra for (A) PTAA, (B) p-FTAA, (C) p-KTAA, and (D) h-FTAA. The lines above and left to the heat maps indicate if a specific spectrum is associated with mCWD (green) or mSS (orange). For all the dyes the emission profiles are separated into 2 distinct clusters corresponding to mCWD (green) or mSS (orange) deposits.
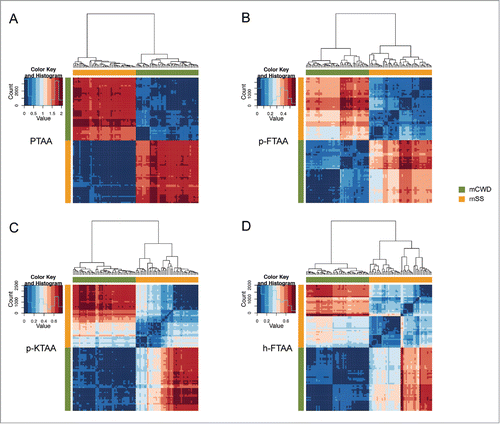
Figure 4. Heat maps and dendrograms for Euclidean distances between pairs of excitation spectra from 50 different regions from protein deposits associated with the respective prion strain. Rows and columns in the heat maps are ordered according to hierarchical clustering of the spectra for (A) PTAA, (B) p-FTAA, (C) p-KTAA, and (D) h-FTAA. The lines above and left to the heat maps indicate if a specific spectrum is associated with mCWD (green) or mSS (orange). For all the dyes the excitation profiles are separated into 2 distinct clusters corresponding to mCWD (green) or mSS (orange) deposits. For h-FTAA (D) the excitation profiles displayed some partial overlap.
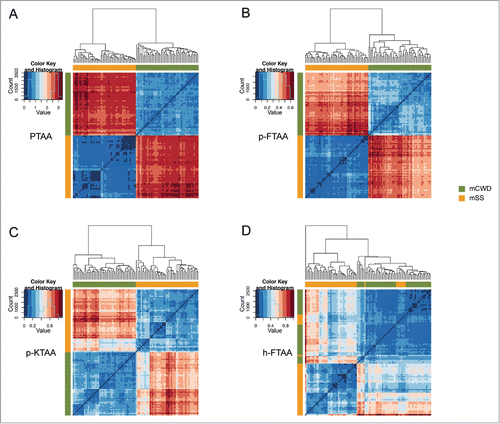
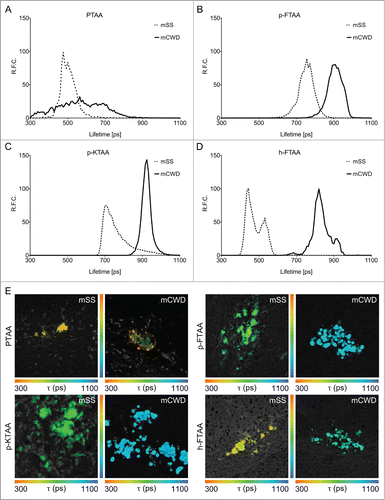
Figure 5. Fluorescence lifetime imaging of thiophene based amyloid ligands bound to PrP deposits. Lifetime decay curves of (A) PTAA, (B) p-FTAA, (C) p-KTAA, and (D) h-FTAA bound to mCWD (black line) and mSS (dotted line) deposits. (E) Fluorescence lifetime images of LCP and LCO stained mCWD and mSS deposits in brain tissue section. The color bars represent lifetimes from 300 ps (orange) to 1100 ps (blue) and the images are color coded according to the representative lifetime. The fluorescence lifetimes were collected with excitation at 490 nm.