Figures & data
Figure 1 Congo red binding with amyloid fibrils and bacterial inclusion bodies. Congo red staining under bright field (left) and showing birefringence under cross-polarized light (right) when binding with amyloid fibrils of HET-s (upper) and inclusion bodies of mouse prion protein PrP(23–231) (lower). (Partial reproduction of ,Citation26 and S8,Citation52).
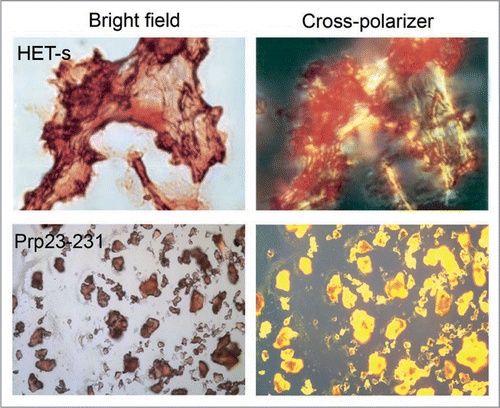
Figure 2 Morphology of bacterial inclusion bodies observed under electron microscope. (A) An E. coli cell containing inclusion body of ESAT-6, (B) Inclusion bodies of BMP2(13–74) after purification, (C) Inclusion body of BMP2(13–74) after incubation at 37°C for 12 hours, (D) Inclusion body of BMP2(13–74) after grown in host cells for 12 hours, (E) Inclusion body of BMP2(13–74) after disaggregated in 4 M of urea solution, (F) Inclusion body of ESAT-6 after incubation at room temperature for 14 days, (G) Inclusion body of Aβ42 after proteinase K proteolytic action, (H) Inclusion body of HET-s(218–289) after three hours expression. Scale bars indicate 1 µm. (Partial reproduction of , S7,Citation52 ,Citation53 and S4,Citation46).
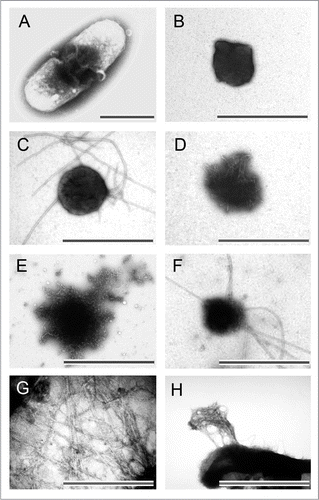
Figure 3 (A) The FTIR spectrum of inclusion bodies of VP1LAC (continuous line) and soluble VP1LAC (broken line). VP1LAC stays in the soluble cell fraction as well as in inclusion bodies when produced in E. coli. (B) The X-ray diffraction of inclusion bodies of ESAT-6. (Partial reproduction of Figs. 8,Citation39 and ,Citation52).
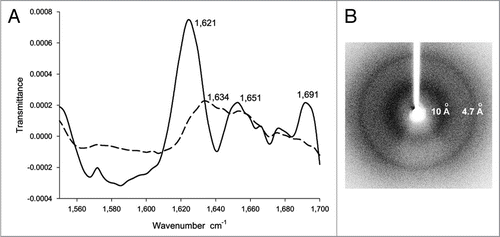
Figure 4 Residues 7–23 of ESAT-6 form cross-β structure in inclusion bodies (A) [15N,1H]-HMQC-spectra of dissolved monomeric ESAT-6 inclusion bodies before (left) and after (right) exchanged in D2O buffer for 311 hours. (B) Ribbon representation of the 3D structure of soluble ESAT-6, while in complex with CFP-10 (not shown).Citation64 The green-colored segment corresponds to residues 7–23. (C) NMR H/D-exchange curves for residues A9, T23, K57 and S77 of ESAT-6. The peak intensities for each residue were plotted versus the H/D exchange time. (D) H/D-exchange rates kex/h, and the relative population P of the two exchange components against the amino acid sequence of ESAT-6. The exchange rates of the major population (p > 1/2) are colored green. If the minor population (p < 1/2) is present more than 1/3, the corresponding exchange rates are shown in grey. The secondary structures of the soluble conformation shown in (B) are highlighted in red. (Partial reproduction of and S2,Citation52).
![Figure 4 Residues 7–23 of ESAT-6 form cross-β structure in inclusion bodies (A) [15N,1H]-HMQC-spectra of dissolved monomeric ESAT-6 inclusion bodies before (left) and after (right) exchanged in D2O buffer for 311 hours. (B) Ribbon representation of the 3D structure of soluble ESAT-6, while in complex with CFP-10 (not shown).Citation64 The green-colored segment corresponds to residues 7–23. (C) NMR H/D-exchange curves for residues A9, T23, K57 and S77 of ESAT-6. The peak intensities for each residue were plotted versus the H/D exchange time. (D) H/D-exchange rates kex/h, and the relative population P of the two exchange components against the amino acid sequence of ESAT-6. The exchange rates of the major population (p > 1/2) are colored green. If the minor population (p < 1/2) is present more than 1/3, the corresponding exchange rates are shown in grey. The secondary structures of the soluble conformation shown in (B) are highlighted in red. (Partial reproduction of Figs. 2 and S2,Citation52).](/cms/asset/71fce615-13d7-4013-a145-b967d9aa73cd/kprn_a_10909922_f0004.gif)
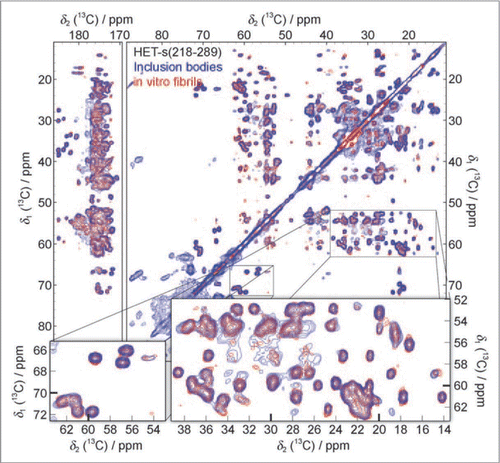
Figure 5 13C-13C solid-state NMR correlation spectrum of purified HET-s(218–289) inclusion bodies (blue) compared to a spectrum of in vitro fibrillized HET-s(218–289) (red). Additional cross-peaks, not belonging to HET-s(218–289), at ∼34 ppm, ∼60–100 ppm and ∼176–180 ppm are assigned to phospholipids, other proteins and RNA from E. coli. (Reproduction of ,Citation46).