Figures & data
Figure 1. Schematic illustration of SP-PLA. (A) Captured antibodies are immobilized on paramagnetic beads. (B) When the sample is incubated with the beads, the targeted PrPs are captured. (C) The same clone of antibody is combined with streptavidin-DNA oligonucleotide conjugates to create PLA probes that are incubated with the captured targets. (D) Oligonucleotides attached to antibodies that have bound in close proximity to each other are joined by enzymatic ligation in the presence of a connector DNA oligonucleotide. (E) Only ligated oligonucleotide reporter molecules can be amplified, detected, and quantified by real-time quantitative PCR.
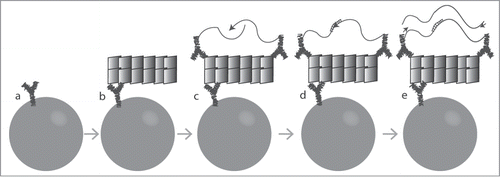
Figure 2. Effect of denaturation of hamster brain homogenates with guanidine-HCl (Gnd-HCl) prior to SP-PLA. (A) A homogenate from an infected hamster brain was incubated with different concentrations of Gnd-HCl and with a control without Gnd-HCl. Dilution series of the homogenates were analyzed for PrPSc through SP-PLA using the monoclonal antibody 3F4. (B) The effect of Gnd-HCl was also examined for a 100-fold dilution of infected and uninfected brain homogenate. Plotted are the mean cycle numbers at threshold (Ct) with standard deviations from duplicate experiments.
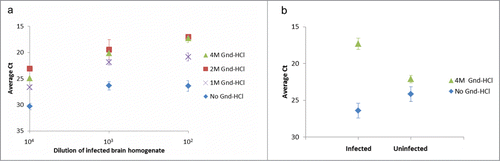
Figure 3. Distinguishing PrPSc aggregates in brain homogenate from an infected hamster vs. an uninfected control hamster. Dilutions of homogenates from brains of infected or uninfected hamsters in buffer were analyzed for the presence of PrPSc using SP-PLA with the 3F4 monoclonal antibody. A cut-off value was set at 3 standard deviations above the mean signal from a negative control (only buffer). Plotted in the graph are the ratios of reporter DNA molecules in the assayed samples relative to this cut off. The data shown are the mean values of 3 replicates with standard deviations.
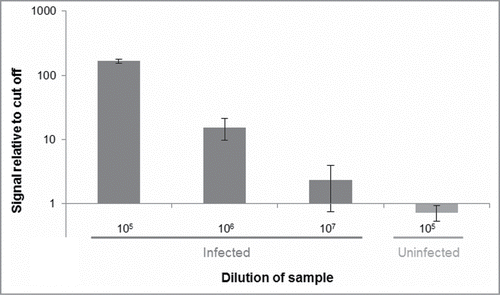
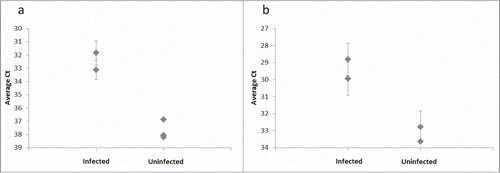
Figure 4. Distinguishing PrPSc aggregates in brain homogenate from several infected and uninfected hamsters. (A) The 3F4 SP-PLA was used to detect PrPSc aggregates in brain homogenates from 2 infected hamsters and from 3 uninfected hamsters diluted 10 million times. (B) An SP-PLA based on the monoclonal antibody 6H4 was used to detect PrPSc aggregates in the same samples diluted 1 million times. Each sample was assayed in duplicates. Plotted are the mean cycle numbers at threshold (Ct) with standard deviations.