Figures & data
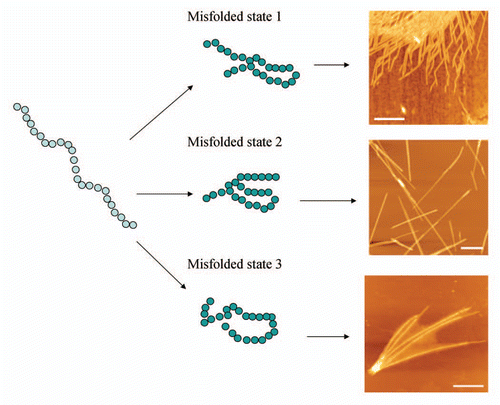
Figure 1 Scheme illustrating that a protein molecule can adopt several conformational states which may result in aggregates of different morphologies. AFM images of CGNNQQNY peptide from Sup35 yeast prion protein aggregated with the formation of fibrils of distinct morphologies at different conditions. Scale bar is 500 nm.