Figures & data
Figure 1 Western blot of the cell extracts after PrPC induction & infection. (A) iN2a cells were cultured for three days with increasing amounts of doxycycline. Cells were lysed at 80% of confluence and equal amounts of protein were analyzed by western blotting for PrPC using the antibody SAF32. (B) iN2a cultured for three passages with 200 ng/mL of doxycycline (iN2a+) were infected with 22L brain prion homogenate as detailed in the Materials and Methods section. After passage 3 or 5, control (Ctrl) and infected cells (22L) were analyzed for PrPC and PrPSc expression (PK− and PK+). Infected iN2a+ cells (iN2a+22L) accumulated significant amounts of PrPSc.
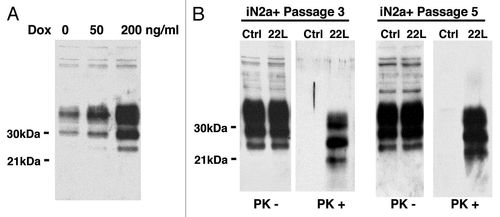
Figure 2 2DE reference gel of iN2a cells. iN2a cell extract were separated using 2DE electrophoresis in a dry strip pH 3–10 for the first dimension, a 12% SDS-PAGE for the second dimension and silver stained. The identification of proteins, noted with their ID number (Suppl. Tables 1 and 2), was performed by peptide mass fingerprints after trypsin digestion and MALDI-TOF on Coomassie or silver stained spots.
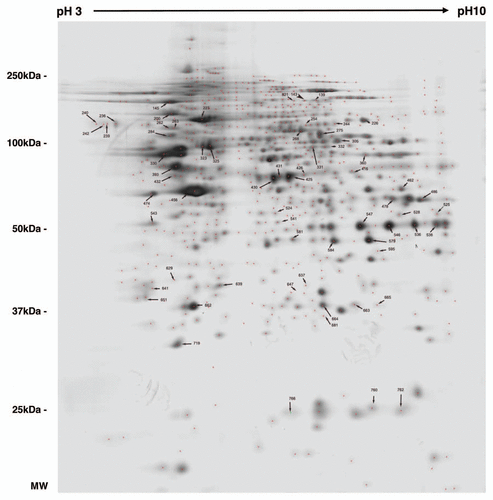
Figure 3 Distribution of the identified proteins per functional blocks (A) and cellular origin (B). (References in Suppl. Table 2.)
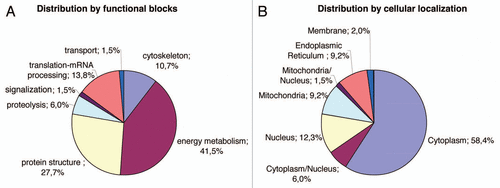
Figure 4 Illustration of protein differential expression. (A) Regions of 2DE silver stained gel of the different cell types illustrating the variation of calreticulin, ATP synthase, lactate dehydrogenase and voltage-dependent anion-selective channel protein. (B) Western blot detection after 1D gel electrophoresis of PDIA6, tubulin beta and vimentin in iN2a, iN2a+ and iN2a+22L. Numbers in parenthesis refer to protein ID numbers (Suppl. Tables 1 and 3).
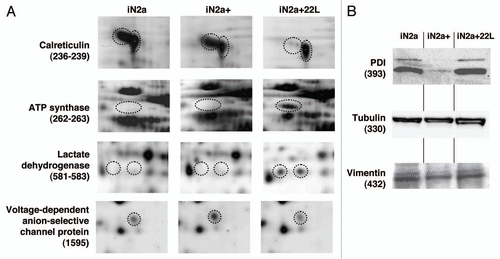
Table 1 Table of identified proteins (spot ID, gene, accession number from UniprotKB database, name and functional group) sorted in eight groups (A-H) based on the fold expression ratio iN2a+/iN2a and iN2a+22L/iN2a+