Figures & data
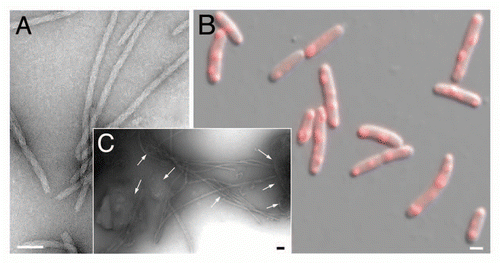
Figure 1 The bacterial RepA-WH1 prionoid highlighted at the molecular and cellular levels. The stable, soluble dimers of the isolated RepA-WH1 domain (dWH1, 1HKQ.pdb) undergo a conformational transformation upon transient, low affinity (albeit sequence-specific) binding to dsDNA ligand, thus resulting in metastable, aggregation-prone monomers (mWH1, modeled on 1REP.pdb). The roughly invariant (rms deviation for 76 Cα atoms: 1.34 Å) WH fold is depicted in cyan, whereas segments showing a significant structural shift are shown in blue. The amyloidogenic peptide L26VLCAVSLI34 (C-terminal 2/3 of α2),Citation21 is colored in red, with the side-chain of the hyper-amyloidogenic mutant residue V31 (bold) rendered as CPK spheres. A possible pathway for the exposure of the amyloidogenic stretch is sketched (dashed grey arrows 1–3): (1) DNA binding to dWH1 results in a bend in the β-hairpin (β2–β3),Citation17 which carries the sensor Arg residues (R78, R81, R91 and R93; yellow),Citation22 thus disrupting the dimerization interface (second dWH1 subunit in grey). (2) Simultaneously, β2–β3 pushes the N-terminal α1 that swingsCitation16 and partially unfolds. (3) The opening of such α1 latch releases C-terminal α5 which, in the whole mRepA, will then pack with the N-terminus of the WH2 domain to build an antiparallel β-sheet,Citation17 but in the isolated mWH1 is unfolded (dashed coil) to leave the amyloidogenic stretch largely exposed to the solvent and ready to refold and assemble into an amyloid cross-β sheet.Citation21 Models rendered with PyMOL (www.pymol.org).
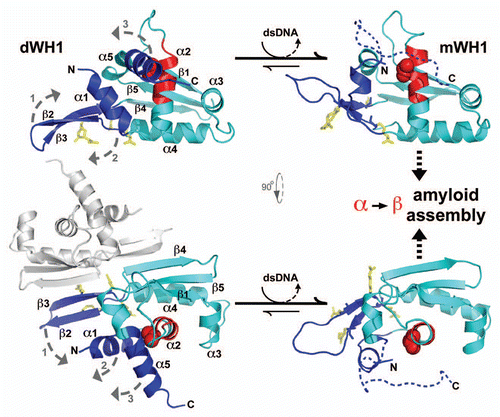
Figure 2 Linking propagation of the bacterial RepA-WH1 prionoid in vitro and in vivo. (A) Electron micrograph of amyloid fibers assembled in vitro by RepA-WH1(A31V) in the presence of effector dsDNA molecules.Citation21,Citation22 Magnification bar: 0.1 µm. (B) E. coli K-12 expressing mCherry-tagged RepA-WH1(A31V) accumulate multiple red fluorescent, aggregated amyloid foci which severely hamper cell proliferation.Citation25 Magnification bar: 1 µm. (C) Molecular transmissibility, the ability of purified bacterial amyloid inclusions/seeds (arrows) to template the transformation and assembly into fibers of soluble RepA-WH1(A31V) molecules,Citation25 links together amyloidosis in vitro and in vivo. Magnification bar: 0.1 µm.