Figures & data
Figure 1 Schematic features of miR cassettes and knockdown efficacy in N2a cells. (A) Each miR cassette in pcDNA6.2-GW/EmGFP vector. (B) Western blot of N2a cells transfected with each miR expression construct. (C) Quantitation of PrPC expression. When miRdual was introduced into the cells, PrPC expression was the most efficiently decreased as compared with wild-type cells (*p < 0.05, **P <0.01).
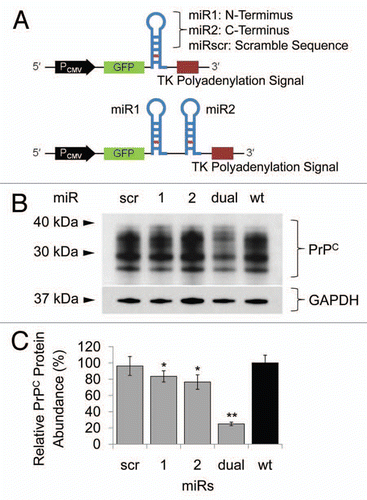
Figure 2 Transfection of miR cassettes and Prnp knockdown efficacy at mRNA and protein level. (A) PCR of genomic DNA shows miR cassette-specific amplicons. Lane L, 100 bp DNA ladder; lane 1, expression vector with miRdual cassette; lane 2, expression vector with miRscr; lane 3, wild-type cells; lane 4, N2amiRdual; lane 5, N2amiRscr; lane 6, no template. (B) qPCR analysis of PrPC transcript abundance showing knockdown efficiency of miRdual in mRNA levels. (C) Western blot analysis of PrPC protein abundance. Top part is the western blot and bottom panel is densitometric evaluation. dual1 and dual2 are N2amiRdual cell clones (*p < 0.05, **p < 0.01).
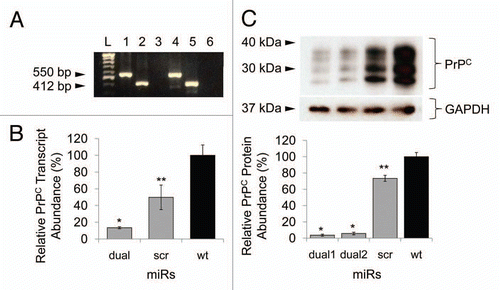
Figure 3 Quantitative PCR analysis of PrPC interacting and/or associated molecules. 6720460F02Rik, Plk3, Ppp2r2b, Csnk2a1 transcripts were upregulated in prion knockdown N2amiRdual cells compared to wild-type N2a cells. The increased Mpg transcripts in both N2amiRdual and N2amiRscr seemed to have no correlation with decreased PrPC (*p < 0.05, **p < 0.01).
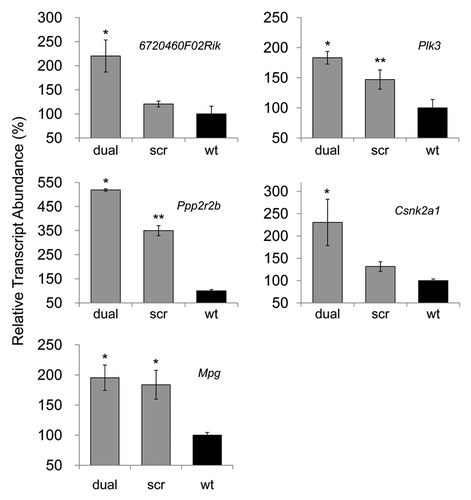
Figure 4 Cell proliferation, viability and GFP expression test. (A) Viability and proliferation of N2a cells stably transformed with miRdual or miRscr (*p < 0.05). (B) Confocal microscopy images of clonal expanded cells. N2amiRdual and N2amiRscr showed green fluorescent signals. Cell nuclei were stained with DAPI (blue). Photos were taken at 20 doubling passages. GFP signals stably expressed over 135 doubling passages. Scale bar, 50 um.
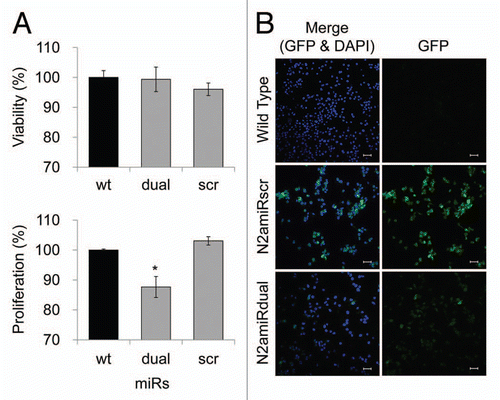
Figure 5 Apoptotic resistance to serum deprivation. Cells were cultured under conditions of serum deprivation and collected at indicated time points. (A) Detection of Drp1 expression levels analyzed by densitometric analysis of western blot. (B) Cytochrome C expression in cytosolic fractions detected by immunoassay. (C) Analysis of caspase 3 activity calculated by comparison with the free pNA. dual, N2amiRdual; scr, N2amiRscr; wt, miR non-treated wild-type N2a (*p < 0.05, **p < 0.01).
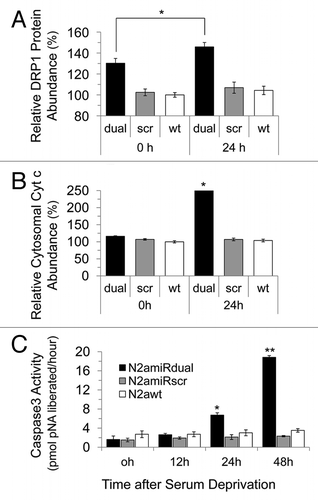
Figure 6 Detection of apoptotic cells following serum deprivation using Annexin V-Cy3 assay. Cells were collected at the indicated time points and 5 × 105 cell suspensions were treated with Annexin V. The proportion of cells showing apoptotic change was increased in N2amiRdual whereas most of cells showed the resistance against apoptosis stimulation in N2amiRscr and wild-type N2a cells (*p < 0.05).
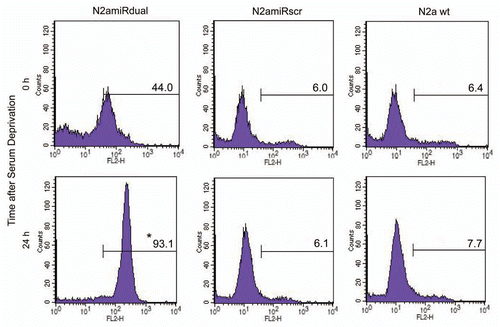
Figure 7 Downregulation pattern of PrPC in RK13-moPrnp cells harvested at different confluences. RK13-moPrnp cells were lentiviral-transduced with miR expression cassettes and the relative abundances of PrPC and GFP processed were determined by western blot (*p < 0.05, **p < 0.01).
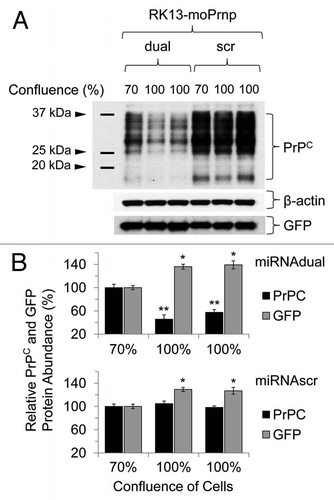
Table 1 Target genes of qPCR for characterizing the established N2amiRdual cell line
Table 2 Sequences of artificial miRs targeting Mus musculus Prnp ORF
Table 3 Primer sequences used in this study