Figures & data
Figure 1 Neuropathology and PrPcore characteristics of L-BSE-affected TgHaNSE mice. (A) hematoxylin and eosin staining of the corpus callosum of mice. (B) PrPSc deposition was detected in the semiserial sections. PrP-plaque is indicated by an arrow. PrP was detected by mAb SAF-84. Scale bars: 200 mm. (C) PrPSc in L-BSE affected TgHaNSE mice was detected by western blotting. Lane 1: C-BSE (cattle), lane 2: L-BSE (cattle), lane 3: C-BSE affected hamster, lane 4: L-BSE affected hamster, lanes 5 and 6: L-BSE affected TgHaNSE, lane 7: scrapie Obihiro-affected mouse. PrP was detected by mAb T2.
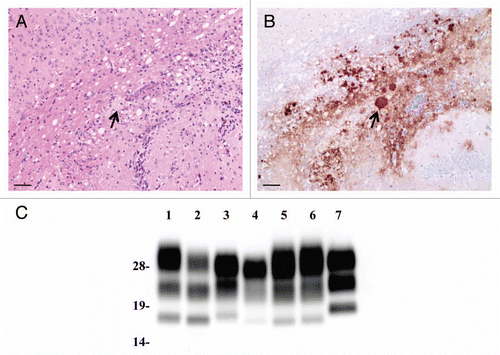
Figure 2 Neuropathological lesion profiling and PrPSc deposition in hamsters. (A) Vacuolar lesion profiles in Syrian hamster brains as observed for scrapie strains Sc237, C-BSE-affected hamsters (C-BSE) and L-BSE-affected hamsters (L-BSE). Gray matter scoring areas: 1, dorsal medulla; 2, cerebellar cortex; 3, superior colliculus; 4, hypothalamus; 5, medial thalamus; 6, hippocampus; 7, septum; 8, posterior cerebral cortex; 9, anterior cerebral cortex. White matter scoring areas: 1*, cerebellar white matter; 2*, midbrain white matter; 3*, cerebral peduncle. Mean (standard deviation) (n = 4). (B) PrPSc deposition in the brains of hamsters affected with C-BSE (second passage), L-BSE (second passage) and Sc237 (serial passage). MAb SAF84 was used for immunostaining. Upper, cerebral cortex; lower, cerebellum.
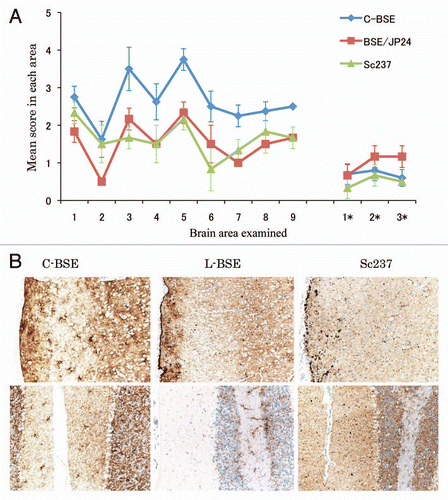
Figure 3 Western blotting analysis of PrPcore from C-BSE- and L-BSE-affected hamsters. (A) Lane 1: C-BSE-affected cattle (natural case); lane 2: L-BSE-affected cattle (natural case); lanes 3 and 6: Sc237-affected hamsters; lanes 4 and 7: C-BSE-affected hamsters; lanes 5 and 8: L-BSE-affected hamsters; lanes 6–8: PNGaseF treatment. (B) The relative amount (%) of di-, mono- and non-glycosylated PrPcore. The results are the mean (standard deviation) of three experiments. Bar diagram: di- (black), mono-(grey) and nonglycosylated form (white).
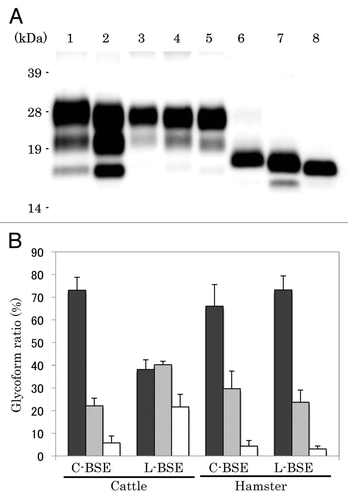
Figure 4 Relative PK resistance of PrPcore in prion-affected hamsters. (A) Western blot results. Lanes 1–4: hamster-adapted C-BSE; lanes 5–8: L-BSE-affected hamster. Lane 9: mouse scrapie prion. The samples were treated with 40 (lanes 1 and 5), 100 (lanes 2 and 6), 500 (lanes 3 and 7) and 1,000 (lanes 4 and 8) µg/ml of PK at 37°C for 1 h. PrPcore was detected with mAb 6H4. Molecular markers are shown on the left (kDa). (B) Relative amount (%) of PrPcore after different PK concentration were indicated. Black circle: C-BSE affected hamster, black square: L-BSE affected hamster, white circle: C-BSE affected cattle, white square: L-BSE affected cattle. Cattle results are obtained from previous study in reference Citation11.
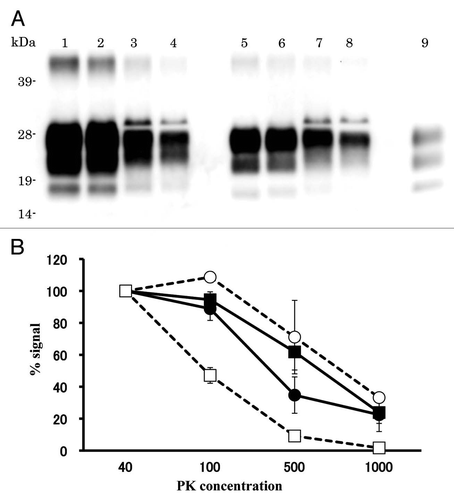
Table 1 Transmissibility of L-BSE in mice
Table 2 Incubation period of hamsters inoculated with C-BSE and L-BSE
Table 3 Comparison of the PrP amino acid sequences in mouse and hamster