Figures & data
Figure 1 pH effect on the oligomerization pattern of V136 R154 Q171 (sheep numbering) PrP sheep variant from size-exclusion chromatography (after heating for 30 min 80 µM of protein at 50°C), following the methods already described in reference Citation11. At pH below 3.5 (blue line) the amount of O3 oligomers increases whereas at pH 4.1 (black line) and 7.15 (red line) the O1 oligomers are the most representative object. At pH 4.5 (green line) no oligomers were detected.
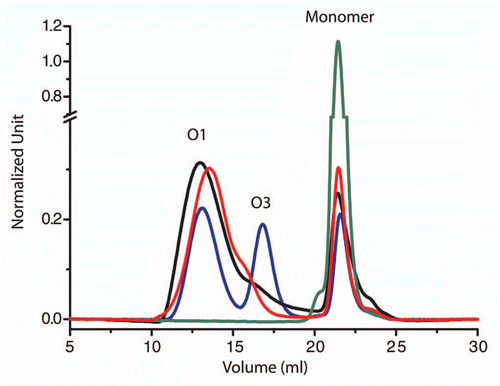
Figure 2 Effect of structural constraint on ovine PrP by additional disulfide bridge on oligomerization process (after heating for 90 min 80 µM of protein at 50°C). (A) Localization of the SS bounds on PrP structure. Left: Y160C M209C, right: V169C E224C (sheep numbering). (B) Their effect on the oligomerization process (black curves: wild-type ovine PrP (ARQ sheep variant); red curves: Y160C M209C and V169C E224C).
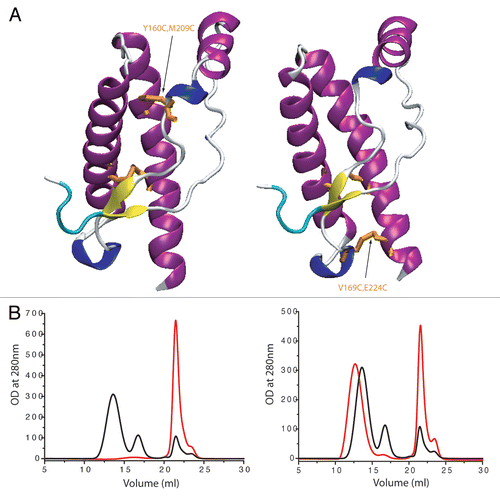
Figure 3 Prion structural dynamics in relation with polymerization process: mechanism of separation of S1H1S2 from H2H3. According to experiments with artificial disulfide bridge, H/D exchange and the fact that H2H3 summarizes the oligomerization pattern of full-length PrP, the activated state prior to aggregation corresponds to the separation of S1H1S2 from H2H3.
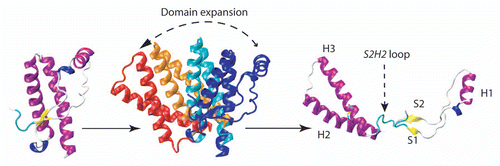
Figure 4 The structural basis of PrPC-to- PrPSc conversion according to S1S2 seeding model. The two β-strands S1 and S2 were proposed to play the role of “seed” for an elongation during the H1 unfolding.Citation44
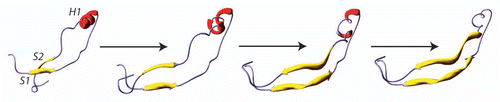
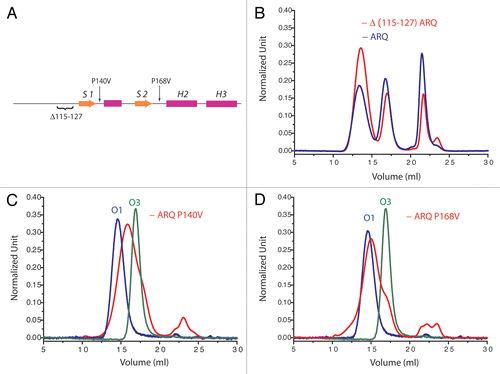
Figure 5 Deletion and mutations in N-terminal and S1H1S2 region affects oligomerization pattern. (A) The localization of the deletion and mutations on PrP structure. Size-exclusion chromatographs of (B) D(115–127) ARQ, (C) ARQ 140V and (D) ARQ P168V (sheep numbering). On chromatographs (C and D) is indicated the position of purified O1 (blue line) and the one of O3 oligomers (green line) from ARQ wild type. These observations indicate that mutations on S1H1S2 affect also the oligomerization pathway.