Figures & data
Figure 1 The cellular environment modulates prion misfolding pathways to create protein-based traits. (top) Self-replicating protein conformations create stable phenotypes (white colonies) in vivo when the processes of synthesis (1), conversion (2), fragmentation (3) by Hsp104 (hexamer) and transmission (4) are balanced to allow aggregates of prion proteins to persist in vivo. (middle) Overexpression of a prion protein promotes the conversion reaction (red arrows), leading to the accumulation of large aggregates that are inefficiently transmitted to daughter cells (dotted red arrow) and loss of the prion-associated phenotype (red colonies). (bottom) Dominant inhibition of prion propagation by mutants that decrease conversion efficiency (dotted red arrow) or enhance fragmentation efficiency (solid red arrow) promote aggregate disassembly (ball and stick) and induce prion loss (red colonies).
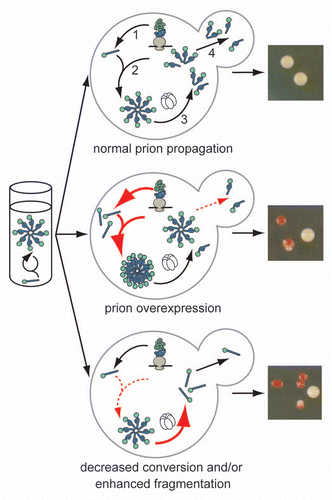
Figure 2 Relating prion phenotypic severity to aggregate thermodynamic stability. For many prion variants, there is a linear but inverse relationship between aggregate thermodynamic stability and phenotypic severity (black dotted line), but this trend cannot explain the phenotypes associated with all prion variants or the effects of dominant-negative prion mutants (see text for details). Our studies in vivo on the yeast prion [PSI+] suggest that thermodynamic stability poses a limit on prion persistence at both extremes (red line) by impacting aggregate size and accumulation (shown schematically). The least thermodynamically stable aggregates are efficiently resolubilized, while the most thermodynamically stable aggregates are inefficiently transmitted.
![Figure 2 Relating prion phenotypic severity to aggregate thermodynamic stability. For many prion variants, there is a linear but inverse relationship between aggregate thermodynamic stability and phenotypic severity (black dotted line), but this trend cannot explain the phenotypes associated with all prion variants or the effects of dominant-negative prion mutants (see text for details). Our studies in vivo on the yeast prion [PSI+] suggest that thermodynamic stability poses a limit on prion persistence at both extremes (red line) by impacting aggregate size and accumulation (shown schematically). The least thermodynamically stable aggregates are efficiently resolubilized, while the most thermodynamically stable aggregates are inefficiently transmitted.](/cms/asset/56823a06-0c44-444e-9b76-72fec2b5b72a/kprn_a_10916413_f0002.gif)
Figure 3 Transmission of Sup35 protein to daughter cells is conformation-dependent. (left) Schematic of fluorescence loss in photobleaching assay (FLIP) for Sup35 transmission to daughter cells. Bleached daughter (red) and monitored mother (black) are indicated. (right) Fluorescence retention in mother cells expressing Sup35-GFP in the [PSI+]strong (white), [PSI+]weak (pink) or [psi−] (red) conformation.
![Figure 3 Transmission of Sup35 protein to daughter cells is conformation-dependent. (left) Schematic of fluorescence loss in photobleaching assay (FLIP) for Sup35 transmission to daughter cells. Bleached daughter (red) and monitored mother (black) are indicated. (right) Fluorescence retention in mother cells expressing Sup35-GFP in the [PSI+]strong (white), [PSI+]weak (pink) or [psi−] (red) conformation.](/cms/asset/5ef91a15-3bc9-44c0-b8c9-92c0bc548768/kprn_a_10916413_f0003.gif)